
ДНК-нанотехнологии 1 введение и основные методы / 10.1021@acs.chemrev.0c00294
.pdf
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 19. DNA origami strategy for the construction of wireframe DNA nanostructures. (A) 2D arrays with cavities, hexagonal patterns and S-shaped structures formed from gridiron units. On the right was the AFM image of S-shaped structures. Adapted with permission from ref 286. Copyright 2013, American Association for the Advancement of Science. (B) Multiple-layered crossovers and corresponding sca old folding pathways, whose AFM images were shown on the right. Adapted with permission from ref 68. Copyright 2016, Wiley-VCH. (C) Complex DNA wireframe structures with four-armed branched DNA, such as flower-and-bird patterns (as shown in the AFM image). Adapted with permission from ref 67. Copyright 2015, Springer Nature. (D) Wireframe DNA origami nanostructures with flexible gap region transformed into rigid DNA nanostructures via DNA polymerase-assisted gap-filling reaction. Adapted with permission from ref 70. Copyright 2018, American Chemical Society.
into branched DNA and linkers via traditional solid-phase synthesis due to the structural similarity, and further selfassembled into nanogel by tile-mediated assembly.276,277 By advantage of the intrinsic cytotoxic of nucleoside analogs, DNA nanohydrogels had antitumor property. In addition, Ge-rich strands exhibited the same behavior as i-motif, that is, folding into intramolecular quadruplex structures at acidic environment, thus endowing the DNA nanohydrogels with pH-responsive- ness. This feature accelerated the disassembly of nanogel and drug release in response to endosomal environments, facilitating enhanced anticancer therapy. On the basis of DNA self-assembly strategy, a method of host−guest interactions was proposed to participate in the formation of nanohydrogel (Figure 18C).278 The multivalent branches had the linking sites of host (β- cyclodextrin) and guest (adamantly grafted PEG polymer), providing convenience for the multidirectional connectivity in DNA nanogel. The introduction of hydrophobic network exhibited a high hydrophobic drug loading capacity. In addition, the size was tuned from micro to macro scale depending on the concentrations of the host and guest molecules. The size adjustability and excellent biocompatibility made DNA nanohydrogel a promising nanocarrier for good cell permeability, intracellular imaging, e cient drug delivery, and enhanced gene therapy.279
Single-stranded tile (SST) was a representation of tilemediated assembly that only one DNA strand was used as building units to complete the assembly of well-defined structures.280 The one strand was designed to contain di erent domains, each one having self-complementary palindromic sequence. Through the intermolecular cross-linking of among individual strands, DNA hydrogel eventually produced.281−283 SST strategy validated the limiting situation of minimum requirement for the number of strands to assemble into DNA materials, fully demonstrating the potential of tile-mediated strategy in assembly.284 SST method greatly reduced the assembly cost and operation complexity, without strict control of the concentration and metering ration of reactants. Di erent to the traditional method of forming hydrogel, that is, forming motifs from multiple strands at first and then jointing them by sticky ends, SST ensured the formation of DNA hydrogels in a one-pot step.
3.2. DNA Origami
DNA origami was to fold DNA into desired shapes of nanostructures with addressable features. To this end, the long sca old strand was generally used to bind with multiple staple strands. The typical origami was tightly packed in plane with the double-crossover DNA as the local structure. Using branched junctions was not restricted to parallel alignment such that the
U |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |

Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 20. DNA origami strategy for the construction of DNA polyhedrons.65 (A) DNA polyhedron possessing larger molecular weight (more than 5 megadaltons) relative to the conventional size distribution of DNA polyhedrons. (B) Variety of DNA polyhedrons self-assembled from DNA tripods as sti origami tiles in a one-pot annealing. (C) DNA tripods with sti vertex helices and fine-tuned interarm angles assembled into DNA polyhedrons through dynamic connection design. Adapted with permission from ref 65. Copyright 2014, American Association for the Advancement of Science.
Figure 21. DNA origami strategy for the construction of DNA crystals. (A) 3D triangular origami crystals with rhombohedral lattice assembled by the stacking of complementary blunt ends. (B) TEM (scale bar, 100 nm) and SEM (scale bar, 100 and 50 nm inset) of 3D origami crystals. Adapted with permission from ref 289. Copyright 2018, Wiley-VCH.
variegated design for those intricate wireframe structures is reachable.285
Han et al. used four-arm junctions as the building-blocks to construct complex wireframe structures (Figure 19A).286 The branched motif could twist within a torsion angle and the sca old strands could travel in multiple directions. Programmable connections by changing the distance between joints or introducing curvature would produce more complex structures such as 2D, multilayered, and curved gridirons. In addition to planar junctions, the branched motif could also be fabricated in the layered fashion (Figure 19B).68 The DNA strands could travel between layers within predesigned angles to obtain desired geometries. Multilayered DNA structures with controlled framework, therefore, were generated. The branched motif showed flexible and dynamic features in the open design of local structures. To construct wireframe structures with arbitrary
connections, Yan and co-workers proposed the strategy to insert double-crossover junctions to bridge DNA helices, which could be adapted to the construction of 3D architectures (Figure 19C).67 At times, branched motif might bring about the gap regions, which induced the structural collapse of DNA origami. Schmidt and co-workers demonstrated the structural transformation of DNA origami with DNA polymerase filling the gap regions (Figure 19D).70 Upon the polymerization, the filled gaps would help straighten the curved structure due to the relatively rigid double helix.
Branched motif with su cient sti ness was put forward for the larger scale architectures. Some scaling parameters like the edge design would a ect the mechanical sti ness of DNA origami.287 Branched origami was referred to as the branched structural unit made from conventional origami strategy. Branched origami provided rigid vertices and the helix bundles
V |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |

Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 22. Dynamic assembly for the construction of branched DNA-based materials. (A) Dynamic assembly mediated by strand migration and displacement reaction for the construction of DNA dendrimers. Adapted with permission from ref 290. Copyright 2014, American Chemical Society.
(B) Clamped hybridization chain reactions (C-HCR) for the self-assembly of patterned 3D DNA networks. Adapted with permission from ref 292. Copyright 2017, Wiley-VCH.
as the connecting arm were common method to increase the mechanical sti ness.288 Using branched origami as the buildingblocks, larger and more complicated DNA architectures could be constructed.
DNA polyhedra were wireframe architectures that exploit the hierarchical assembly of multiarm-junction motifs. Previous polyhedra were assembled by flexible single-stranded hinges, which hindered the correct assembly of polyhedra with larger sizes and molecular weight (Figure 20A). Iinuma et al. developed a universal strategy to construct polyhedra from megadalton monomers (Figure 20B).65 In terms of structural and functional similarity, the origami motif, or DNA tripods in their work, could be considered as branched origami. To ensure the sti ness of DNA tripods, each arm involved 16 parallel helices and the rigidity of vertices could be strengthened through vertex helices. The DNA tripods had controlled interarm angles and di erent connecting manner would result in distinct assembly of polyhedral nanostructures (Figure 20C). Despite the challenges such as low reaction kinetics, the massive branched origami possessed powerful capability for the construction into higher order structures.
Crystals were orderly arranged microscopic structures and the formation of DNA crystals requires higher spatial accuracy. The lack of periodic symmetry of DNA origami, however, was unfavorable for the growth of lattices. In the work by Liedl, the tensegrity triangle motif was built out of DNA origami (Figure 21A).289 The branched motif had three struts in an overunder fashion and each strut contained 14 helix bundles. Because of the existence of seam in the helix bundles, the growth of the triangle motif was triggered in the presence of the polymerization strands via the blunt-end stacking (Figure 21B). Importantly, such rigid branched origami also allowed for the variable localization of guest molecules.
3.3. Dynamic Assembly
In addition to using repetitive elements to construct DNA dendrimers, another DNA dynamic assembly based on strand displacement reaction was also applied for the formation of DNA dendrimers. As shown in Figure 22A, the assembly process required two dsDNA as substrates, two ssDNA as assistants and a trigger input.290 Substrates were designed to include an
exposed toehold for the activation by a trigger chain, an internal sequence for the hybridization with corresponding assistant, and bulge loops that were complementary to the toehold domain in another substrate. First, substrate-A was dissociated by the combination of trigger input and assistant-A, resulting in byproduct-A and intermediate. Two bulge loop sequences were exposed in the intermediate, and further served as the trigger chains for substrate-B. With the help of assistant-B, two substrate-B were dissociated and bound to the intermediate, resulting in branched DNA with exposed triggers. The branched DNA was in turn used as trigger input for the dissociation of substrate-A. After several rounds of cycling, a spontaneous assembly process for the formation of DNA dendrimers was completed. A cascade of strand displacement reactions was initiated by target-specific triggering, while the substrates remained intact in the absence of target. This feature greatly simplified the operation complexity and enabled the process to be completed in a one-pot manner. Another prominent feature is that the target-triggered method achieved the exponential growth of DNA dendrimers; as a consequence, the dynamic assembly had vast potential in e ective signal amplification, molecule sensing, and clinical diagnostics.291
Clamped hybridization chain reaction (C-HCR) is a typical strand displacement reaction-based dynamic assembly strategy to construct high-level structures. A clamped core in the C-HCR system worked in a way similar to seeds for crystal growth, enabling the bridging and locking the DNA branches (Figure 22B).292 Like traditional HCR, three DNA strands including two hairpin strands and an initiator strand were used in this system. Particularly, one of the hairpin strands had ten palindromic bases; thus the dimer of the hairpin strand had two branches, which could form a three-arm junction or a fourarm junction for divergent chain reactions. With the help of DNA initiators, downstream hybridization and cross-linking occurred and led to expanded and clamped 3D networks, namely all-DNA hydrogels. This dynamic construction strategy facilitated in situ encapsulation of cells in hydrogels and toeholdmediated responsive release.293
On the basis of the isothermal strand displacement in the tensegrity triangle units, a three-state DNA device was built that
W |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 23. Strand displacement reaction driving the conformation changes of DNA nanostructures. (A) Substitution of fluorescence dyes-labeled strands in 3D crystals device inducing the three-stated color changes. Adapted with permission from ref 69. Copyright 2017, Springer Nature. (B) Conformation transition of DNA nanocages by toehold-mediated strand displacement process. Adapted with permission from ref 225. Copyright 2015, Wiley-VCH.
Figure 24. pH-responsive reversible assembly of branched DNA-based materials. (A) pH-responsive hydrogel assembled by the Y-shaped DNA units with three interlocking domains. Adapted with permission from ref 61. Copyright 2009, Wiley-VCH. (B) Reversible assembly and disassembly of DNA tetrahedron with triplex-cohesion in response to pH of solution. Adapted with permission from ref 295. Copyright 2013, American Chemical Society.
could be operated within the context of a self-assembled 3D DNA crystal (Figure 23A).69 The device consisted of an extension of the central strand of the triangle, which was operated by inserting and removing the strands based on singlestranded branch migration. As a result, the transitions between three states can be observed from color changes within the crystal like a tra c light. Through the clever design of sequences and the assistance of strand displacement reactions, dynamic 3D structural and topological transformation between di erent types of DNA polyhedrons could be accompanied (Figure 23B).225 This strategy provided a way to assemble complex DNA nanocages that cannot be assembled from their component tiles directly, in which the dramatic conformational changes were not easily predicted. In addition, owing to inherent wireframe structure, DNA tetrahedrons had a large surface porosity, which could be reversibly switched by aid of strand hybridization and displacement process.127
The introduction of stimuli-responsive elements enabled dynamic assembly and changes in the morphology and topology of DNA-based materials. In 2009, Liu and co-workers reported a fast, pH-responsive and pure DNA hydrogel through the selfassembly of Y-shaped DNA (Figure 24A).61 The Y-shaped DNA containing intermolecular i-motif domains was used to interlock neighboring Y-units, which endowed the rapid gel-to-sol transition of DNA hydrogel with pH responsiveness and allowed for the controlled capture and release of gold nanoparticles. In replace of i-motif domain, two triplex structures including protonated cytosine-guanine-cytosine (C- G·C+) bridge and thymine-adenine-thymine (T-A·T) bridge, which were embedded in the sticky ends of Y-shaped DNA, also made pure DNA hydrogels undergo reversible assembly and disassembly processes upon the activation of specific pH values.294 The introduction of responsive elements also enabled the reversible assembly and disassembly of DNA tetrahedrons
X |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |

Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 25. Dynamic assembly for the construction of DNA arrays. (A) ATP-triggered responsive assembly of DNA ladder by advantage of aptamerligand binding. (B) Nε-p-nosyl-L-lysine (PSL)-triggered assembly of DNA ladder. Adapted with permission from ref 302. Copyright 2019, American Chemical Society.
Figure 26. Branched DNA-gold nanoparticles (AuNPs) hybrids. (A) DNA-AuNPs hybrid dendrimers in controlled spatial arrangement. TEM images of corresponding DNA-AuNPs (Inset). Adapted with permission from ref 324. Copyright 2016, Royal Society of Chemistry. (B) Target-induced AuNP dimers with Y-shaped DNA duplex. (C) Y-shaped DNA-AuNP dimers as a colorimetric sensor for the detection of target DNA with di erent concentrations. Adapted with permission from ref 325. Copyright 2013, American Chemical Society.
under specific environments. For example, the incorporation of |
DNA and dsDNA linkers, they introduced a kind of polymer |
cytosine-containing triplex allowed the formation or dissocia- |
polypropylene oxide (PPO) with thermal-responsiveness in situ |
tion of tetrahedrons in response to pH changes (Figure 24B).217 |
capsuled to the self-assembled DNA network.299 When the |
Since base pairing is one of the most important driving forces |
temperature was lower than the lower critical solution |
for DNA self-assembly, the process of assembly can be |
temperature (LCST), PPO was hydrophilic and evenly |
dynamically regulated responding to thermal by influencing |
dispersed in 3D DNA network, namely the solution states; |
base pairing. In 2011, Liu and co-workers further demonstrated |
when the temperature was in the range of LCST and the melting |
a new assembly method to prepare DNA hydrogel.296 The DNA |
temperature of DNA network, PPO became hydrophobic and |
hydrogel was self-assembled by using Y-shaped DNA as sca olds |
self-collapsing at the single molecular level, which induced the |
and dsDNA as linkers, possessing the gel-to-sol transition |
transformation of DNA network to DNA hydrogel. Relative to |
properties in response to thermal. In addition, the conformation |
single-network hydrogel, double-network or double-cross-link- |
changes of DNA linkers enabled the DNA hydrogels to undergo |
ing hydrogel can o er more flexible operation and more |
a gel-to-sol phase change and enhanced mechanical strength of |
complicated stimulus responsiveness.300,301 |
DNA hydrogel.297 The mechanical performances (such as |
A general approach aptamer-based, allosteric mechanism for |
elastic and viscous modulus) of the DNA hydrogel showed |
diversified stimuli responsive DNA self-assembly was further |
temperature-reversible percolation transition, which depended |
developed (Figure 25).302 The response to stimuli relied on a |
on the melting temperature of sticky ends and the flexibility of |
designable DNA motif with DNA aptamer incorporated. In the |
linkers.298 On the basis of DNA pure hydrogel consisting of Y- |
presence of the cognate ligand such as ATP and Nε-p-nosyl-L- |
Y |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |

Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 27. Branched DNA-based regular structures for the ordered assembly of AuNPs. (A) Oriented AuNPs arrangement with DNA arrays as templates through attaching two kinds of AuNPs (5 and 10 nm) on di erent three-point-star branched DNA. TEM of 2D AuNPs arrays was reproduced with permission from ref 328. Copyright 2006, American Chemical Society. (B) Schematic patterning and corresponding fast Fourier transform (FFT) image of AuNPs occupied in cavities of hexagonal DNA lattices. Adapted with permission from ref 329. Copyright 2019, American Chemical Society. (C) Schematic diagram of the formation of methane-like AuNPs using branched DNA-based tetrahedron as a fixed framework. (D) TEM image of methane-like AuNPs (10 nm diameters), and corresponding cryoEM images and 2D projections derived from reconstructed computer model. Adapted with permission from ref 77. Copyright 2015, American Chemical Society. (E) Hybrid assembly of a library of polyhedron DNAAuNPs frameworks. Adapted with permission from ref 78. Copyright 2016, Springer Nature.
lysine (PSL), the aptamer bound to the ligand and causes the DNA motif to change from a flexible interior loop to a stable, rigid helical conformation. The single-stranded overhang base pair with the bulges to form T-junctions, which can further assemble into large nanostructures. The platform established by this strategy could be applied to allow for the DNA self-assembly to respond to other stimuli.
3.4. Hybrid Assembly
Branched DNA could form hybrid materials with functional moieties including inorganic nanoparticles (e.g., gold nanoparticles,303 silver nanoclusters,304 magnetic nanoparticles,305 and quantum dots79), small organic molecules (e.g., polyaniline,306 cholesterol,307 and fluorescent dyes308), organic molecules-transition metal complex,85 and biomacromolecules (e.g., peptides309 and proteins310). These novel hybrid DNA materials combined the advantages of both branched DNA and functional moieties.311
3.4.1.DNA-Inorganic Nanoparticles Hybrids.
3.4.1.1.DNA-Gold Nanoparticles Hybrids. Gold nanoparticles (AuNPs) have become the object of long-standing fascination
owing to intense surface plasmon resonance, size-related electronic, magnetic, and optical characteristics.312 The aggregation of AuNPs induced by salt concentrations was easy to occur, causing the color changes due to the coupling of
neighboring plasmons and the changing of resonance frequency.313 The method of DNA-grafting provided a new
idea for preventing the uncontrolled aggregation of AuNPs and maintaining colloidal stability.25,314−316 To accurately control the spatial arrangement and ordered assembly of AuNPs, a particularly appealing strategy was to use tailorable DNA
nanostructures as permanent sca olds or templates to custom the morphology and framework of AuNP nanoassemblies with site-specific addressability.317−322 Branched DNA, as a welldefined nanostructure, provided a powerful way for building ordered and shaped-directed nanoparticles architectures owing to its precise configuration, controlled nanoscale and multifunctionality.323 Inspired by thiol-gold conjugation, a large number of highly organized materials consisting of thiolmodified DNA and AuNPs have been developed.25,26 Chen et al. reported a hyperbranched self-assembled nanostructure based on DNA dendrimers and AuNPs (Figure 26A).324 The sticky ends of DNA dendrimer were distributed in a controlled fashion around the periphery of the 3D structure, which served as a sca old to place AuNPs in precise spatial positions. Guo et al. constructed a colorimetric sensor based on target-induced and controlled AuNPs aggregation (Figure 26B).325 After sequential PEG modification and probe immobilization, asymmetric AuNPs with dense PEG shell was obtained, which did not aggregate in the absence of target DNA. In contrast, when in the presence of target DNA, Y-shaped DNA was formed to minimize the interparticle distance of asymmetrically PEGylated AuNPs, resulting in shoulder-by-shoulder AuNP dimers, which led to a significantly enhanced stability and detection sensitivity (Figure 26C).
To achieve more precise spatial organization of AuNPs, 2D DNA array and 3D DNA polyhedron DNA have overincreasingly emerged as attractive templates.78,326,327 Zheng et al. fabricated a regular AuNP array attached on preassembled 2D rhombic lattice from DNA tensegrity triangle.328 The motif contained cohesive ends elongated from two linear independent directions to form lattice skeleton, and a third direction as an
Z |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
Chemical Reviews |
pubs.acs.org/CR |
Review |
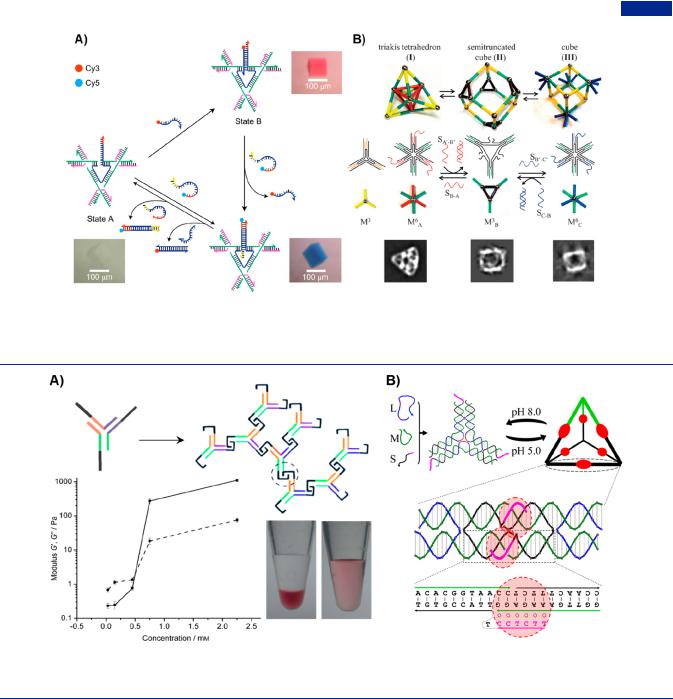
Figure 28. Branched DNA-silver nanoparticles (AgNPs) or silver nanoclusters (AgNCs) hybrids. (A) Construction of super-AgNC with three di erent types of branched DNA as templates, which possessed excellent tunable fluoresent properties. Adapted with permission from ref 80. Copyright 2018, Wiley-VCH. (B) Ag+-cross-linked DNA hydrogel undergoing sol-to-gel transition by the addition and removal of Ag ions. Adapted with permission from ref 332. Copyright 2014, Royal Society of Chemistry. (C) AgNCs formed by the sequence-specific Y-shaped DNA as templates exhibiting red-emitting or yellow-emitting fluorescence properties. Adapted with permission from ref 333. Copyright 2013, Wiley-VCH.
addressable position to link AuNP, finally producing periodic patterns of AuNPs with di erent diameter (Figure 27A). The ordered AuNP array could also be formed via the surface adsorption of AuNP and substrate where anchored DNA array served as structural molds, rather than the covalent linkage of AuNPs to modified DNA.329 DNA templates with uniform cavity patterns first attached on the substrate surface (such as mica) driven by the base stacking of bridged and blunt-ended DNA motifs. The substrate surface was coated by positively charged materials as “glues” for strong trapping of AuNP. The DNA array-templated AuNPs patterns with high cavity occupancy were eventually obtained (Figure 27B). This strategy was not limited to the integrity and homogeneity of template, and the diversity of NPs. In addition, He et al. reported a DNAtemplated strategy for the formation of 2D metallic NPs arrays in the gas−solid phase.330 The organization of AuNPs patterns was dictated by DNA template, and the size of AuNPs was controlled by the amount of gold during thermal deposition and evaporation coating, finally obtaining 2D AuNPs arrays with controllable topography. Li et al. reported molecule-analogous NP clusters by encapsulating AuNPs into DNA framework nanocages.77 The DNA nanocages with customized shape and orientation served as guiding agents for the assembly of NPs with defined valences and directional bonds, such as DNA tetrahedron for CH4-like NP-molecule (Figure 27C,D), DNA cube for SF6-like NP-molecule and DNA bipyriamid for W(CH3)6-like NP-molecule. In addition, the AuNPs guest could be further released from the AuNP@DNA core−shell nanocages in a control and predetermined matter through ingenious strand displacement reactions.331 The DNA-capped AuNPs could also serve as adhesive to realize the particle-frame
assembly from a large diversity of polyhedrons (Figure 27E).78 The resulting NPs-DNA frameworks exhibit high-ordered lattice and crystallographic symmetry in accordance with the geometry of DNA polyhedron frames, The approach facilitated the desired organization and rational assembly of nanoscale particles.
3.4.1.2. DNA-Silver Nanoparticles Hybrids. Silver nanoclusters (AgNCs) have been widely applied in promising biomedical fields such as bioimaging, biosensing, and antibacterial. Generally, the preparation of AgNCs was applied by template methods, and the choice of template had an important influence on the properties of AgNCs such as fluorescence, catalysis, and antibacterial. Latorre et al. prepared a family of DNA-AgNCs stabilized with benzene ring-centered branched DNA, whose fluorescence intensity and stability were significantly improved compared with ssDNA likely due to the vicinal e ect.304 Yang et al. reported a series of branched DNA nanostructures as sca olds to form superbranched silver nanoclusters (super-AgNCs), which possessed tunable spatial structures and fluorescence properties (Figure 28A).80 They systemically studied the e ects of di erent molecular structures and arm lengths on optical performance of super-AgNCs. Interestingly, super-AgNCs exhibited better antibacterial properties than AgNCs formed by the ssDNA template. In addition, super-AgNCs showed excellent biocompatibility and low biotoxicity, which enabled adaptivity in a variety of biomedical applications. Willner group reported an Ag+-cross- linked reversible DNA hydrogel and a fluorescent DNA hydrogel by incorporating AgNCs into DNA hydrogel
containing Y-shaped DNA as building units (Figure 28B,C).332,333 The Ag+-cross-linked gelation depended on the
formation of cytosine-Ag+-cytosine bridges in duplex regions
AA |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
Chemical Reviews |
pubs.acs.org/CR |
Review |
between Y-shaped DNA and dsDNA linkers. Furthermore, the removal of Ag+ ions could induce the dissociation of DNA hydrogel under the action of cysteamine ligand (Figure 28B). In addition, AgNCs stabilized in the sequence-specific loop domain of Y-shaped DNA exhibited red-emitting or yellow-emitting fluorescence properties, endowing the DNA hydrogel with fluorescence tunability and thermo-controllable gel-to-sol switching (Figure 28C).
3.4.1.3.DNA-Magnetic Nanoparticles Hybrids. Magnetic nanoparticles (MNPs) have been applied in a myriad of fields due to their inherent magnetic e ects, including magnetic
resonance imaging, magnetic hyperthermia therapy and biosensing.334 Ma et al. introduced DNA-modified MNPs, Y- sca olds and DNA linkers into the frame of DNA hydrogels, so
as to utilize magnetic field to control the certain shape and desirable direction of DNA hydrogels.76 It is noteworthy that the chemically covalent bonds between amino-MNPs and thiolated ssDNA could improve the stability of hybrid DNA-MNPs hydrogels. In addition, the DNA-MNPs hydrogels could undergo gel-to-sol transition in response to temperature, enzyme and magnetic field. Wang et al. designed titanium
dioxide (TiO2)-coated magnetite microspheres-DNA dendrimers bioconjugates as signal amplifiers for the electro-
chemical detection of polynucleotide kinase activity.335 With the help of polynucleotide kinase and ATP, 5′ end of DNA sequences was activated to produce phosphorylated DNA
capture probes, which were specifically modified on the TiO2- coated MNPs to generate DNA-TMNP dendrimers. In the presence of DNA link probes, the DNA-TMNP dendrimers functioned for e cient signal amplification were attached on the gold electrode. Because the MNPs with peroxidase-like activity could catalyze the reduction of hydrogen peroxide, the
synergistic e ect of MNPs, TiO2, and DNA dendrimers generated remarkable electrochemical signal for the sensitive detection of polynucleotide kinase activity.
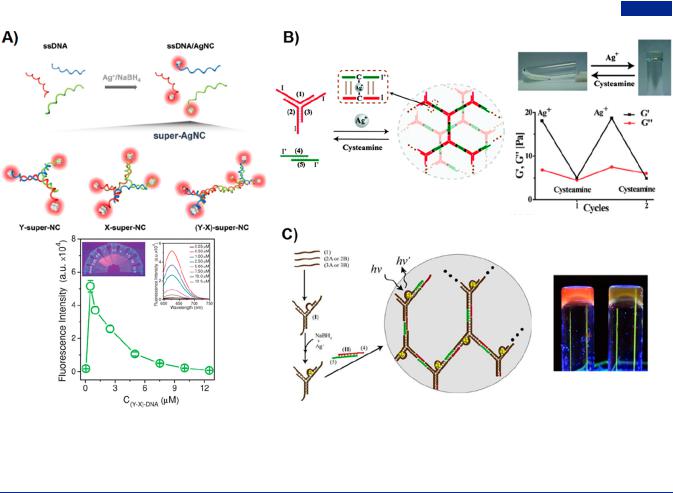
3.4.1.4.DNA-Quantum Dots Hybrids. QDs were attractive in many fields such as materials engineering, biosensing and in vivo imaging due to the fluorescent tunability, solubility, photo-
bleaching resistance, long-term stability, and other prominent characteristics.336−338 The introduction of QDs endowed branched DNA-based materials with more functionality. In 2017, Zhang et al. created a series of DNA-QDs hybrid hydrogels by the one-step self-assembled hybridization of QDs
functionalized ssDNA as linkers and Y-shaped DNA (Figure 29A).79 The QDs-DNA hybrid hydrogels showed size and spectral tunability by regulating the reaction time (Figure 29B). In addition, the QDs-DNA hybrid hydrogels possessed high quantum yield, low photobleaching, and long-term photostability. With the loading of drug and the incorporation of aptamer, the multifunctional DNA hybrid nanohydrogels were used as nanocarriers for cell-specific targeting and enzymecontrolled drug delivery, and further showed increased therapeutic e cacy. The DNA hybrid nanohydrogels were potentially applied in intracellular imaging. Shen et al. reported
engineering QDs of customized valency by employing programmable branched DNA as sca olds.339 The QDs were first
modified with mercaptopropionic acid (MPA) to improve aqueous solubility, and then capped by neutral mPEG to regulate surface charge for facilitating the approaching of DNA (Figure 29C). The MPA/mPEG cocovered QDs were incubated with the di erent number of chimeric DNA strands to prepare engineering-valent DNA-QDs hybrids. The chimeric DNA strands included two portions: phosphonothioate DNA
Figure 29. Branched DNA-quantum dots (QDs) hybrids. (A) Fabrication of a series of DNAQD hybrid hydrogels from ssDNA linkers and Y-shaped DNA. (B) Tunable fluorescence properties of DNA-QD hybrid hydrogels by regulating reaction time.79 (C) Preparation diagram of monovalent DNA-QD hybrid by incubating MPA/mPEG cocoated QDs and phosphonothioate DNA (ptDNA) with the help of steric exclusion and electrostatic repulsion. (D) Customized valency of ptDNA-QD hybrids self-assembled from chimeric DNA strands, including divalent, trivalent and tetravalent QDs and QDs-AuNPs with 5 and 10 nm diameter, Adapted with permission from ref 339. Copyright 2017, Wiley-VCH.
(ptDNA) as nanocrystal ligands and phosphate portion as recognition sequences. Furthermore, the ptDNA standing up to 50 bp provided steric hindrance so that DNA-QDs was selfassembled by both steric exclusion and electrostatic repulsion strategies. The multivalent DNA-QDs hybrids had the potential for the bottom-to-up fabrication of heterodimers (Figure 29D).
3.4.2. DNA-Small Organic Molecules Hybrids. Apart from branched DNA labeled with nanoparticles, the hybrid conjugation of DNA and small organic molecules could be diverse and tunable.44 Representative examples included hydrophobic moieties-DNA, namely amphiphilic DNA and fluorophore-DNA conjugates.81,340 These additional molecules could achieve embedded functionality other than branched DNA and further enrich the underlying potentials in nanoelectronics, nanophotonics and biomedical areas.177,308
Amphiphilic molecules tended to support highly ordered structures, which have been utilized to construct well-defined
AB |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
Chemical Reviews |
pubs.acs.org/CR |
Review |
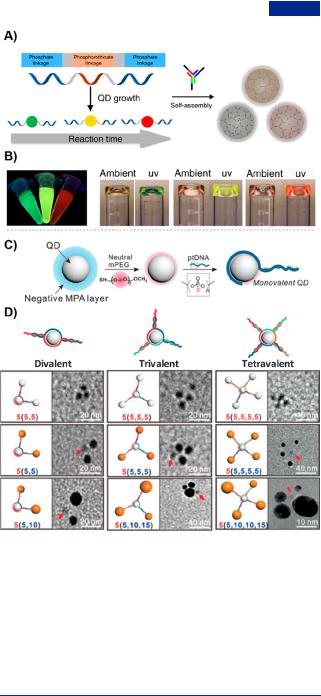
Figure 30. Amphiphilic branched DNA-cholesterol hybrids. (A) Design of amphiphilic branched DNA and the construction of 3D DNA crystal by hydrophobicity-mediated self-assembly. Diverse design included well-defined arm length, specific chemical modification and embedded functionality. Optical image was reprinted with permission from ref 307. Copyright 2017, American Chemical Society. (B) Bright field images, 2D di raction patterns and radially averaged profiles of rhombic-dodecahedral DNA crystal with the increasing of arm length. (C) Partition of di erent macromolecular samples in amphiphilic DNA crystal with di erent arm length (bp). Scale bar, 10 μm. Adapted with permission from ref 81. Copyright 2018, American Chemical Society.
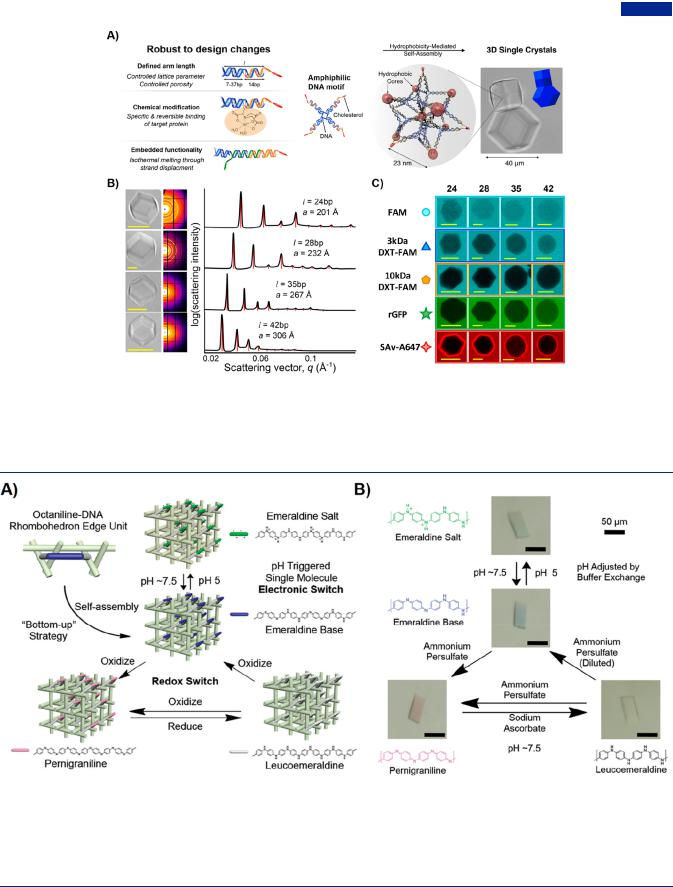
Figure 31. Branched DNA-polyaniline hybrids. (A) Construction of 3D DNA-octameric aniline crystal. The octameric aniline molecule (also called octaniline) as a kind of viable electronic switch, exhibited di erent colors in four di erent redox states, which endowed the 3D DNA-octaniline hybrids with unique property of controlled reversible redox conversion. The emeraldine base state of octaniline was converted to a conducting state termed as emeraldine salt at pH 5, and in turn was converted to highest oxidation state (pernigraniline) and fully reduced state (leucoemeraldine) by adding oxidant. (B) Controlled transition of four redox states in a single crystal. The conversion between blue emeraldine base state to green emeraldine salt state was operated by adjusting the bu er pH. The di erent oxidation states were regulated by the addition of reduction or oxidation reagents. Adapted with permission from ref 349. Copyright 2017, Wiley-VCH.
and monodisperse assemblies.341−343 Since pure DNA architectures reliant on Watson−Crick base pairing were easily limited by the increased complexity of DNA assembly, the incorporation of hydrophobic modifications would result in the
formation of amphiphilic DNA motif, which showed unique stability and was expected to avoid the assembly defects.344 Michele group created C-star motif, which was composed of 4 core-forming oligonucleotides as well as 4 cholesterol function-
AC |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 32. Supramolecular assembly for the construction of branched DNA-based hybrids. (A) Transition metal-centered branched DNA as buildingblocks for the guest-mediated construction of defined oligomeric networks and further 1D DNA fibers. Adapted with permission from ref 149. Copyright 2007, American Chemical Society. (B) Metal-DNA hybrid nanocages assembled by the metal-dpp ligands coordination chemistry. Adapted with permission from ref 85. Copyright 2009, Springer Nature. (C) Transition metal-centered branched DNA for the supramolecular assembly of metalated DNA nanotubes. Adapted with permission from ref 350. Copyright 2011, Wiley-VCH.
alized strands. Each nanostar arm included fixed cholesterolized strand (14 bp) and changeable core-forming strand (7−37 bp) (Figure 30A). Slow cooling led to the selfassembly of amphiphilic C-star motif, and resulted in the formation of macroscopic crystals. The increased arm length brought about greater fraction and programmable porosity of the resultant crystal while kept the crystal morphology (Figure 30B). The long-range order was supported by the topology and symmetry of the flexible nanostar motif, and the additional modifications enabled protein entrapment and toeholdmediated strand displacement, indicating the resilience and robustness of the C-star self-assembly (Figure 30C).
Fluorescent dyes could be facilely modified onto DNA sca olds, which promoted the promises of DNA nanotechnology in the field of detection and bioimaging. Synthesized fluorescent probes to target branched DNA probe have been developed.346 Over the years, ongoing e orts have been devoted to the manipulation of the optical energy at the nanoscale level, of which the enhanced transfer e ciency was particularly pursued.347 To this end, programmed DNA assemblies o ered a rigid structural template with directional control over chromophore organization.308 Compared to linear DNA sca olds with poor transfer e ciency, branched DNA e ectively shaped the chromophore orientation, which induced the available light harvesting.340 For example, Buckhout-White et al. assembled more than 550 DNA architectures involving 85 organic dyes network and demonstrated that DNA dendrimers
could achieve complex FRET network with high transfer e ciency.348
Other organic components could also confer electronic and optical performances to branched DNA. One typical example polyaniline was extensively studied as the electrically conducting polymer. Wang et al. incorporated octaniline into DNA tensegrity triangle, one of the rigid DNA branched motif.349 Remarkably, with the building-block self-assembling into the crystal lattice, the redox and optical performances derived from octaniline molecules were preserved (Figure 31A). Whereas newly formed emeraldine base in the neutral bu er was blue, the
protonated emeraldine salt at pH 5 changed to green. On the other hand, by altering the imino/amine ratio through treating with ammonium persulfate, the crystal resulted in the color change from colorless to rose (Figure 31B). Furthermore, branched DNA could act as the template to control the growth of aniline monomers.306
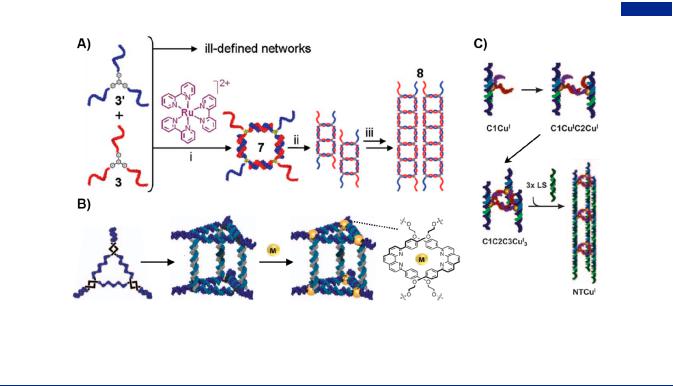
3.4.3. DNA-Organic Molecule/Transition Metal Hybrids. Organic molecule-transition metal complex was acted as the junction vertex to construct branched DNA; in the other way, branched DNA was regarded as the sca old for the precise arrangement of metal complex. The incorporation of transition metal complex eliminated the rigidity of a persistence length of DNA strands, and was amenable to the construction of DNA nanoarchitectures with smaller sizes and improved flexibility. Meanwhile, the chemical linking of transition metal complex and DNA enhanced the thermo-stability of hybrids. In addition, the site-specific distribution of transition metal at the nanoscale endowed the hybrids with the properties of redox, catalysis, magnetism, photochemistry and nanoelectronics. By utilizing transition metal-centered branched DNA, Sleiman group created a class of complex and functional DNA-organic molecule/transition metal hybrids, opening a new area of supramolecular DNA assembly.4,146 The branched strands in DNA-transition metal complex were designed as complementary sequences, which promoted the branched DNA into cyclic oligomeric aggregations by the combined interaction of hybridization and coordination (Figure 32A).149 However, in the absence of transition metal, two branched DNA with organic molecules as center were uncontrollably assembled into illdefined networks, which demonstrated the significant implication of transition metal in supramolecular assembly. The diverse geometry of coordination molecules-branched DNA played a key role in the shape of ultimately formed DNA nanostructures. For example, branched DNA with two strands resulted in a closed ring structure. When the geometry of branched DNA was designed as three-armed branches, the resulting cyclic structure
with exterior strands for the preparation of complex assemblies, such as periodic DNA ladders (Figure 32A).149 The DNA
AD |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
