
- •Preface
- •Acknowledgements
- •Contents
- •List of Contributors
- •1. Array Formats
- •2. Biomolecules and Cells on Surfaces – Fundamental Concepts
- •3. Surfaces and Substrates
- •4. Reagent Jetting Based Deposition Technologies for Array Construction
- •5. Manufacturing of 2-D Arrays by Pin-printing Technologies
- •6. Nanoarrays
- •8. Labels and Detection Methods
- •9. Marker-free Detection on Microarrays
- •13. Analysis of Gene Regulatory Circuits
- •16. Biological Membrane Microarrays
- •18. Whole Cell Microarrays
- •20. Application of Microarray Technologies for Translational Genomics
- •Index


biological and medical physics, biomedical engineering

biological and medical physics, biomedical engineering
The fields of biological and medical physics and biomedical engineering are broad, multidisciplinary and dynamic. They lie at the crossroads of frontier research in physics, biology, chemistry, and medicine. The Biological and Medical Physics, Biomedical Engineering Series is intended to be comprehensive, covering a broad range of topics important to the study of the physical, chemical and biological sciences. Its goal is to provide scientists and engineers with textbooks, monographs, and reference works to address the growing need for information.
Books in the series emphasize established and emergent areas of science including molecular, membrane, and mathematical biophysics; photosynthetic energy harvesting and conversion; information processing; physical principles of genetics; sensory communications; automata networks, neural networks, and cellular automata. Equally important will be coverage of applied aspects of biological and medical physics and biomedical engineering such as molecular electronic components and devices, biosensors, medicine, imaging, physical principles of renewable energy production, advanced prostheses, and environmental control and engineering.
Editor-in-Chief:
Elias Greenbaum, Oak Ridge National Laboratory,
Oak Ridge, Tennessee, USA
Editorial Board:
Masuo Aizawa, Department of Bioengineering,
Tokyo Institute of Technology, Yokohama, Japan
Olaf S. Andersen, Department of Physiology,
Biophysics & Molecular Medicine,
Cornell University, New York, USA
Robert H. Austin, Department of Physics, Princeton University, Princeton, New Jersey, USA
James Barber, Department of Biochemistry,
Imperial College of Science, Technology
and Medicine, London, England
Howard C. Berg, Department of Molecular
and Cellular Biology, Harvard University,
Cambridge, Massachusetts, USA
Victor Bloomf ield, Department of Biochemistry, University of Minnesota, St. Paul, Minnesota, USA
Robert Callender, Department of Biochemistry,
Albert Einstein College of Medicine,
Bronx, New York, USA
Britton Chance, Department of Biochemistry/
Biophysics, University of Pennsylvania,
Philadelphia, Pennsylvania, USA
Steven Chu, Department of Physics,
Stanford University, Stanford, California, USA
Louis J. DeFelice, Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA
Johann Deisenhofer, Howard Hughes Medical
Institute, The University of Texas, Dallas,
Texas, USA
George Feher, Department of Physics,
University of California, San Diego, La Jolla,
California, USA
Hans Frauenfelder, CNLS, MS B258,
Los Alamos National Laboratory, Los Alamos,
New Mexico, USA
Ivar Giaever, Rensselaer Polytechnic Institute,
Troy, New York, USA
Sol M. Gruner, Department of Physics, Princeton University, Princeton, New Jersey, USA
Judith Herzfeld, Department of Chemistry,
Brandeis University, Waltham, Massachusetts, USA
Pierre Joliot, Institute de Biologie Physico-Chimique, Fondation Edmond de Rothschild, Paris, France
Lajos Keszthelyi, Institute of Biophysics, Hungarian Academy of Sciences, Szeged, Hungary
Robert S. Knox, Department of Physics
and Astronomy, University of Rochester, Rochester,
New York, USA
Aaron Lewis, Department of Applied Physics,
Hebrew University, Jerusalem, Israel
Stuart M. Lindsay, Department of Physics
and Astronomy, Arizona State University,
Tempe, Arizona, USA
David Mauzerall, Rockefeller University,
New York, New York, USA
Eugenie V. Mielczarek, Department of Physics
and Astronomy, George Mason University, Fairfax,
Virginia, USA
Markolf Niemz, Klinikum Mannheim,
Mannheim, Germany
V. Adrian Parsegian, Physical Science Laboratory,
National Institutes of Health, Bethesda,
Maryland, USA
Linda S. Powers, NCDMF: Electrical Engineering, Utah State University, Logan, Utah, USA
Earl W. Prohofsky, Department of Physics, Purdue University, West Lafayette, Indiana, USA
Andrew Rubin, Department of Biophysics, Moscow
State University, Moscow, Russia
Michael Seibert, National Renewable Energy
Laboratory, Golden, Colorado, USA
David Thomas, Department of Biochemistry,
University of Minnesota Medical School,
Minneapolis, Minnesota, USA
Samuel J. Williamson, Department of Physics, New York University, New York, New York, USA
U.R. Muller¨ D.V. Nicolau (Eds.)
Microarray Technology
and Its Applications
With 123 Figures
Including 16 Color Plates
123
Uwe R. Muller,¨ Ph.D.
V.P., Applied Science Nanosphere, Inc.
4088 Commercial Avenue Northbrook, IL 60062 USA
e-mail: umuller@nanosphere.us
Prof. Dan V. Nicolau
Swinburne University of Technology 533-545 Burwood Rd.
Hawthorn, Victoria 3122 Australia
e-mail: dnicolau@swin.edu.au
Library of Congress Cataloging in Publication Data: 2004113284
ISSN 1618-7210
ISBN 3-540-22931-0 Springer Berlin Heidelberg New York
This work is subject to copyright. All rights are reserved, whether the whole or part of the material is concerned, specif ically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microf ilm or in any other way, and storage in data banks. Duplication of this publication or parts thereof is permitted only under the provisions of the German Copyright Law of September 9, 1965, in its current version, and permission for use must always be obtained from Springer. Violations are liable to prosecution under the German Copyright Law.
Springer is a part of Springer Science+Business Media
springeronline.com
© Springer-Verlag Berlin Heidelberg 2005
Printed in Germany
The use of general descriptive names, registered names, trademarks, etc. in this publication does not imply, even in the absence of a specif ic statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use.
Cover concept by eStudio Calamar Steinen
Typesetting by the authors using a Springer LATEX-macro package
Cover production: design & production GmbH, Heidelberg
Production: LE-TEX Jelonek, Schmidt & Vöckler GbR, Leipzig
Printed on acid-free paper |
SPIN 10884448 |
57/3141/YL - 5 4 3 2 1 0 |

Preface
It has been stated that our knowledge doubles every 20 years, but that may be an understatement when considering the Life Sciences. A series of discoveries and inventions have propelled our knowledge from the recognition that DNA is the genetic material to a basic molecular understanding of ourselves and the living world around us in less than 50 years. Crucial to this rapid progress was the discovery of the double-helical structure of DNA, which laid the foundation for all hybridization based technologies. The discoveries of restriction enzymes, ligases, polymerases, combined with key innovations in DNA synthesis and sequencing ushered in the era of biotechnology as a new science with profound sociological and economic implications that are likely to have a dominating influence on the development of our society during this century. Given the process by which science builds on prior knowledge, it is perhaps unfair to single out a few inventions and credit them with having contributed most to this avalanche of knowledge. Yet, there are surely some that will be recognized as having had a more profound impact than others, not just in the furthering of our scientific knowledge, but by leveraging commercial applications that provide a tangible return to our society.
The now famous Polymerase Chain Reaction, or PCR, is surely one of those, as it has uniquely catalyzed molecular biology during the past 20 years, and continues to have a significant impact on all areas that involve nucleic acids, ranging from molecular pathology to forensics. Ten years ago microarray technology emerged as a new and powerful tool to study nucleic acid sequences in a highly multiplexed manner, and has since found equally exciting and useful applications in the study of proteins, metabolites, toxins, viruses, whole cells and even tissues. Although still relatively early in its evolution, microarray technology has already superseded PCR technology not only in the breadth of applications, but also in the speed with which this evolution has taken place. Note that the literature dealing with microarrays has increased dramatically from its humble beginnings in the mid-nineties to reach more than 2000 articles and almost 300 reviews in 2004 alone (Fig 1). Although a saturation point may have been reached - not surprisingly given that there is
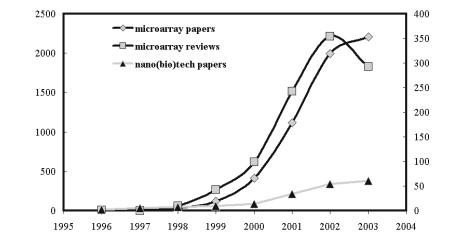
VI Preface
still a limit to the number of laboratories that have access to this technologyits impact is truly remarkable, especially when compared, for example, to the emerging and much touted field of Nanotechnology.
Number of Articles
Reviews of Number
Fig. 1. Comparative evolution of publications regarding microarrays and nanobiotechnology
Amidst the pace of such rapid knowledge expansion, there is a challenge in trying to compose a book that does not face obsolescence by the time of its first publication. Alas, the breadth of this field is driving the growing knowledge base into many new directions, generating the need for di erent books at di erent levels and each with a di erent and unique focus.
As early participants in the development of microarray technology the editors have learned to appreciate the need for contributions from many di erent areas in the basic sciences and engineering that were crucial to its birth and continued healthy growth. In turn we have observed how the involvement in this particular scientific endeavour has a ected many careers, turning physicist into oncologists, physicians into bioinformaticians, and chemists and biologists into optical engineers. Provided the diverse nature of backgrounds that are required to further propel this field, we thought it appropriate to aggregate this book around three aspects of microarray technology: fundamentals, designed to provide a scientific base; fabrication, which describes the current state of the art and compares ‘old’ and new ways of building microarrays; and applications, that are aimed to highlight only the amazing variety and options provided by these techniques. As an aid to the practitioner we have also asked the authors to provide a detailed method section wherever appropriate.
Part 1, General Microarray Technologies, opens with an overview on microarray formats. Chapters 2 and 3 cover the fundamentals of the physicochemical aspects of immobilizing biomolecules on di erent substrates, while
Preface VII
Chaps. 4 and 5 describe the principal techniques used for array manufacture. Chapter 6 explores the limits of miniaturization with nanoarrays, and Chap. 7 illuminates various aspects of microfluidics for automation. Finally, Chaps. 8 and 9 deal with the principles of labelling and detection methodologies. The next parts are concerned with application of these fundamental techniques toward the development and use of specific types of microarrays. Part 2 describes DNA based microarrays in 4 chapters, covering SNP detection, high sensitivity expression profiling, comparative genomic hybridization, and the analysis of regulatory circuits. Part 3 contains 3 chapters that deal with microarrays for protein and small molecule detection, describing array technology for antibodies, aptamers, and lipid bound proteins, respectively. The final part comprises 4 chapters that introduce the most esoteric arrays, those that contain high information content in each feature (whole cells or tissues), and the capability of performing biological reactions, such as transfections. How the combination of these types of arrays generates new insights into the molecular basis of normal and malignant cell function is summarized in the last chapter.
It appears that given the dynamics of microarray technology any book would be a ‘work in progress’. Rather than fighting this, the editors and the authors of this book embrace this concept: chances are that this book will grow in time in line with the new developments in microarray technology.
June, 2004 |
Uwe M¨uller |
|
Dan Nicolau |

Acknowledgements
The initial idea for this book emerged during a serendipitous meeting between the Editors and a representative from Springer Verlag during a Conference on Microarrays, Fundamentals, Fabrication and Applications that was chaired and organized by the Editors as part of the International Society for Optical Engineering (SPIE) Meeting in January 2001 in San Jose, CA. In fact, several Chaps. of this book were authored by people present at that Conference. The Editors wish to thank the organizers of SPIE, and in particular Marilyn Gorsuch and Annie Gerstl, who helped with the organisation of these Conferences in the last four years. Thanks also to the Conference co-Chairs, Ramesh Raghavachari and David Dunn. The Editors also wish to thank Peter Livingston and Gerardin Solana for the tedious work of converting the manuscripts into a camera-ready format.
Many contributors have specific acknowledgements.
The authors of Chap. 1 are grateful to Stephen Felder, Ph.D. and Richard Kris, Ph.D. of NeoGen, LLC. (Tucson, AZ), the inventors of the multiplexed nuclease protection assay, for proof-of-principle work on the mRNA assay and for the software for reagent design, image analysis and data interpretation.
The authors of Chap. 4 would like to thank Innovadyne Technologies for use of Fig. 4.5 and Peter Hoyt for helpful discussions. The research presented here was sponsored by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory (ORNL), managed by UT-Battelle, LLC for the U. S. Department of Energy under Contract No. DE-AC05-00OR22725 and by NIH Grant R01 HL62681-02. The manuscript has been authored by a contractor of the U.S.Government under contract DE-AC05-00OR22725. Accordingly, the U.S. Government retains a nonexclusive, royalty-free license to publish or reproduce the published form of this contribution, or allow others to do so, for U.S. Government purposes.
One of the authors of Chap. 6 (DVN) wishes to thank Dan V. Nicolau Jr. for discussions regarding the computational applications of nanoarrays.
The authors of Chap. 7 would like to thank Joe Bonanno and Dale Ganser (formerly Motorola Labs) for help in device fabrication, and Gary Olsen and
XAcknowledgements
Pankaj Singhal (Motorola Life Sciences) for useful discussions on hybridization kinetics. This work has been sponsored in part by NIST ATP contract #1999011104A and DARPA contract #MDA972-01-3-0001.
Some of the authors of Chap. 8, i.e. JJS and SSM, acknowledge the NIH for support. CAM acknowledges the AFOSR, DARPA, and the NSF for support of this work.
The authors of Chap. 9 are very grateful to Gabriele G¨unther for excellent technical assistance, to Dr. K. B¨ohm, Jena, for kindly providing us with microtubules and kinesin samples, and to Dr. Wolf, PicoRapid GmbH Bremen, for help in spotting protein samples by an automatic arrayer.
The authors of Chap. 13 thank Dr. Tae Hoon Kim and Miss Sara Van Calcar for critical reading of the manuscript. We are also grateful to Drs. Hieu Cam, Yasuhiko Takahashi, Brian Dynlacht, Richard Young, and Mr. Tom Volkert for their help during the development of the technology described in this chapter. B.R. is supported by the Ludwig Institute for Cancer Research and a Sidney Kimmel Foundation for Cancer Research Scholar Award.
The authors of Chap. 14 wish to acknowledge the great support by Dr. Ronald Frank.
Finally, the authors of Chap. 20 thank Juha Kononen, Guido Sauter, Holger Moch, Lukas Bubendorf, Galen Hostetter, Ghadi Salem, John Kakareka and Tom Pohida for their contribution to the tissue microarray development, and Robert Cornelison, Don Weaver, Abdel Elkhahloon, and Natalie Goldberger and for their contributions to the cell microarrays.
