
1Foundation of Mathematical Biology / Foundation of Mathematical Biology
.pdf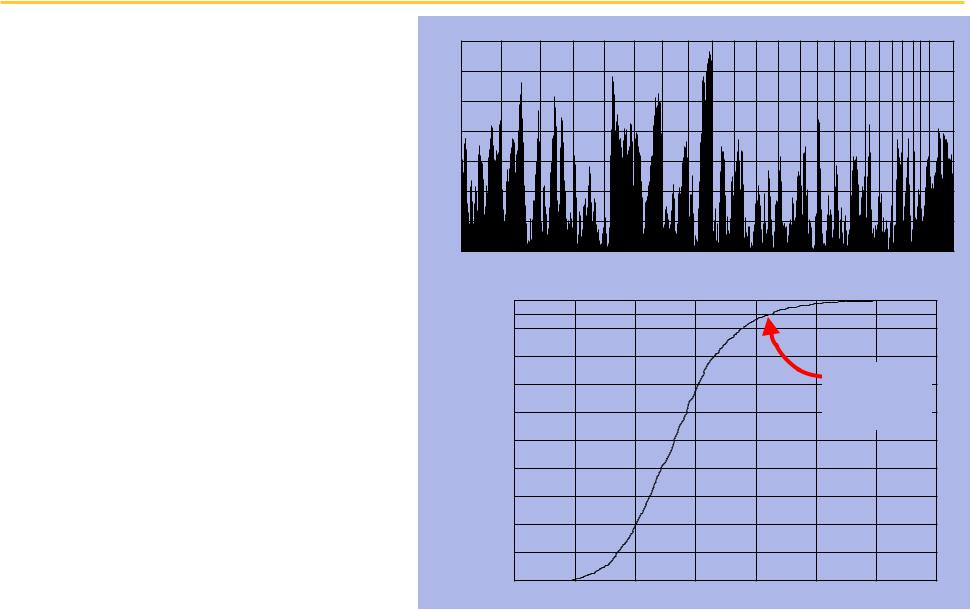
UCSF
Bad news: the correlations appear to be no better than chance at p = 0.05
We compute the direct correlation for each of
1225 loci
♦Strongest correlation at 8q24
♦Many other peaks
Compute level of significance using permutation analysis
We get a critical value of
0.36
Correlation Magnitude
Correlation magnitude with overall survival
0.35
0.3
0.25
0.2
0.15
0.1
0.05
0
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 20 2122 X |
Genomic Position
Cumulative Histogram of Correlation Magnitudes
|
1 |
|
|
|
|
|
|
|
|
0.9 |
|
|
|
|
|
|
|
|
0.8 |
|
|
|
|
|
|
|
|
0.7 |
|
|
|
|
|
p = 0.05 |
|
Proportion |
|
|
|
|
|
|
threshold |
|
0.6 |
|
|
|
|
|
is 0.36 |
|
|
|
|
|
|
|
|
|
||
0.5 |
|
|
|
|
|
|
|
|
Cumulative |
|
|
|
|
|
|
|
|
0.4 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.3 |
|
|
|
|
|
|
|
|
0.2 |
|
|
|
|
|
|
|
|
0.1 |
|
|
|
|
|
|
|
|
0 |
|
|
|
|
|
|
|
|
0.15 |
0.2 |
0.25 |
0.3 |
0.35 |
0.4 |
0.45 |
0.5 |
Correlation Magnitude

UCSF
General Principle: Reduce the number of observations
Any method we can use to subselect a smaller set of observations from the larger set helps us, provided:
♦The subselection method must be orthogonal to the correlation being studied
•If we’re trying to link copy number to survival, we can’t systematically employ the survival outcomes in making our subselection
♦Ideally, the method should have some compelling intuitive support based on the data
♦Restricting observations based on frequency/magnitude is a generally useful technique: it tends to eliminate noise
UCSF
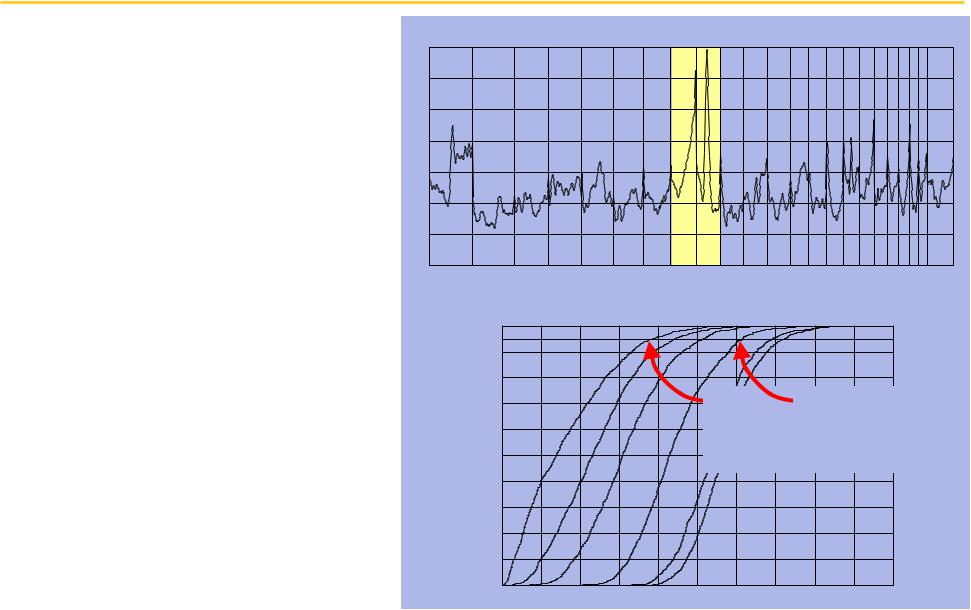
By including frequency and amplitude, we can detect weaker correlations
The magnitude of copy number variation is not uniformly distributed
♦9q13 has the largest cumulative variation
♦8q24 has the next highest
Significance thresholds on correlation vary with
“energy”
♦Energy 0.0, t = 0.36
♦Energy 3.0, t = 0.31
♦Energy 6.0, t = 0.19
|
|
|
|
|
|
|
CGH variation energy |
|
|
|
|
|
|
|
|
|||
7 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
4 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Energy |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 20 2122 X |
|
|
|
|
|
|
|
Genomic Position |
|
|
|
|
|
|
|
|
|
||
|
|
|
Cumulative Histogram of Correlation Magnitudes with Multiple Energies |
|
||||||||||||||
|
|
1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.9 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.8 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.7 |
|
|
|
|
|
|
E = 6.0, |
|
E = 3.0, |
|
|
|||||
|
Proportion |
0.6 |
|
|
|
|
|
|
p = 0.05 |
|
p = 0.05 |
|
|
|||||
|
|
|
|
|
|
|
|
threshold |
|
threshold |
|
|||||||
|
0.5 |
|
|
|
|
|
|
is 0.19 |
|
|
is 0.31 |
|
|
|
||||
|
Cumulative |
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
0.4 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0 |
0.05 |
0.1 |
0.15 |
|
0.2 |
0.25 |
|
0.3 |
0.35 |
|
0.4 |
|
0.45 |
|
0.5 |
|
|
|
|
|
|
|
|
Correlation Magnitude |
|
|
|
|
|
|
|
|
|||
UCSF
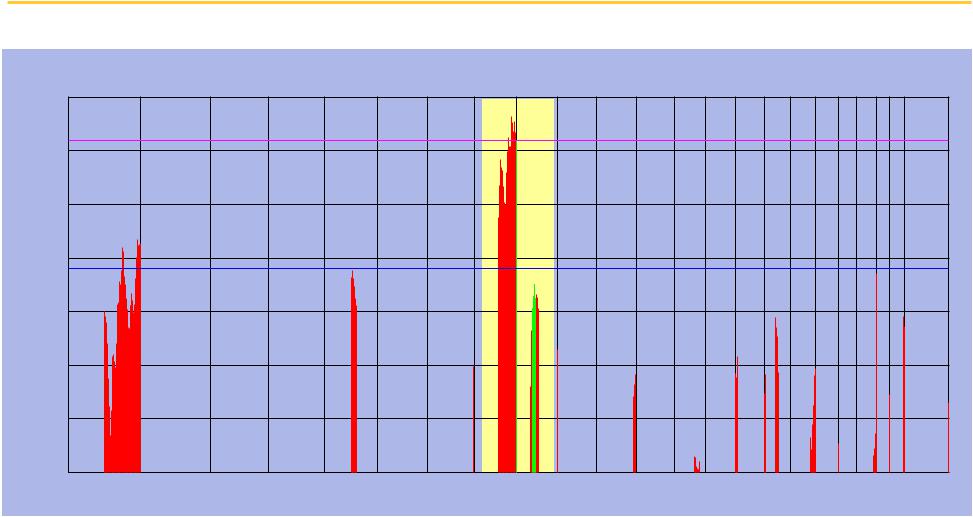
Both 8q24 and 9q13 are significantly correlated with survival
Correlation Magnitude
Correlation Magnitude with Overall Survival
0.35
0.3
0.25
0.2
0.15
0.1
0.05
0
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 20 2122 X |
Genomic Position
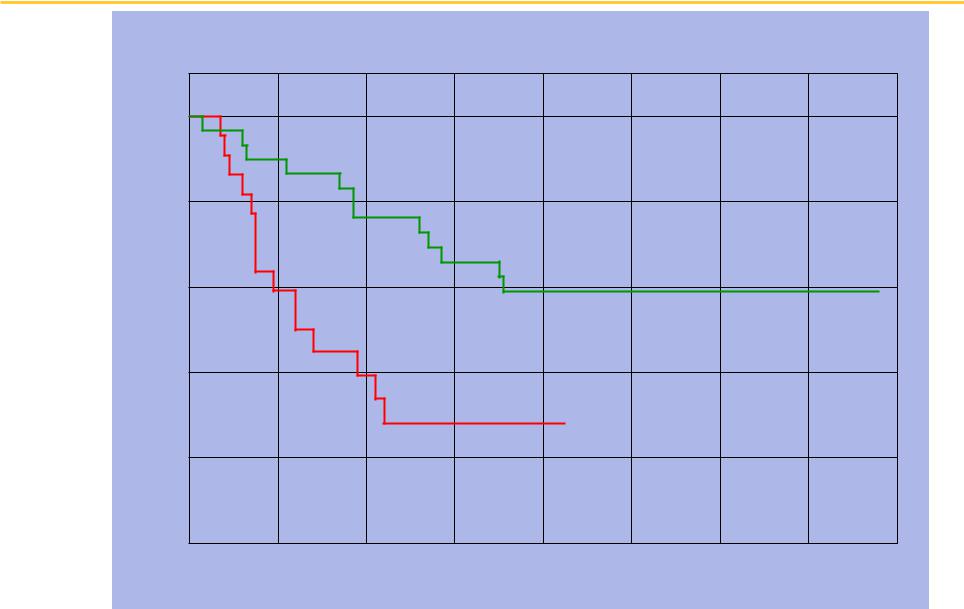
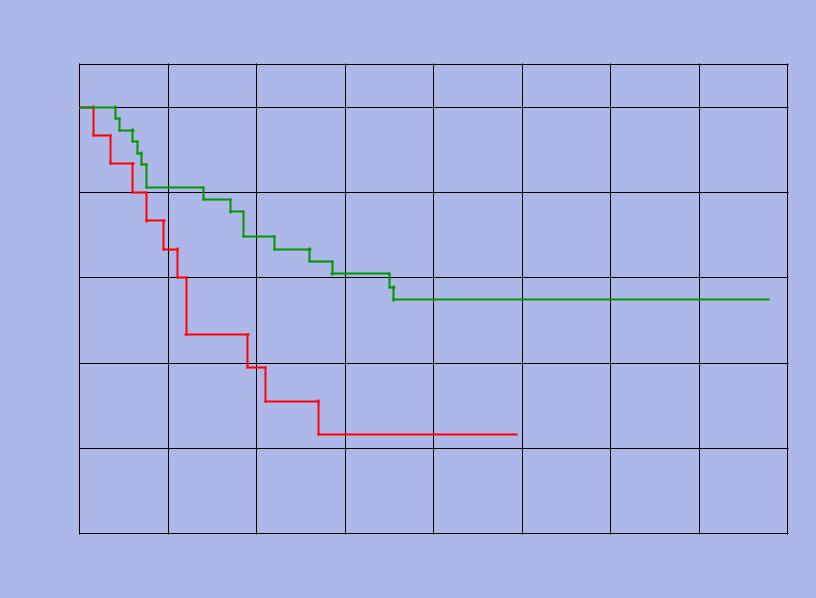
UCSF Amplification at 8q24: poorer survival (p < 0.01)
Kaplan-Meier Plot of Normal vs Amplified at 8q24
Fraction Surviving
1
0.8
Normal
0.6
0.4
Amplified
0.2
0
0 |
20 |
40 |
60 |
80 |
100 |
120 |
140 |
160 |
Survival Duration
UCSF |
Deletion at 9q13: poorer survival (p < 0.01) |
|
|
|
Kaplan-Meier Plot of Normal vs Deleted at 9q13 |
Fraction Surviving
1
0.8
0.6 |
Normal |
|
0.4
Deleted
0.2
0
0 |
20 |
40 |
60 |
80 |
100 |
120 |
140 |
160 |
Survival Duration
UCSF |
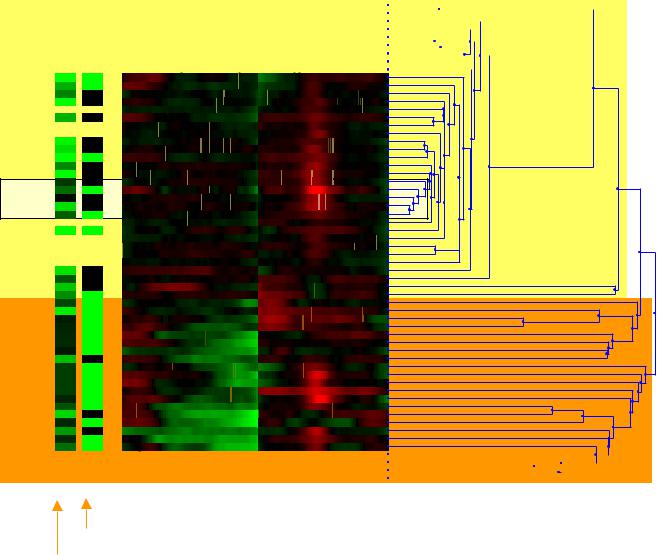
Clustering based on chromosomes 8 and 9 reveal |
|
patterns of survival and tumor phenotype |
||
|
|
|
Cluster profiles based on Chr 8,9
♦Display raw data
♦Display survival, p53 status
Cluster enrichment is statistically significant
♦Orange block
•Surv < 35 months
•p53 often mutant
♦Yellow block
•Surv > 75 months
•p53 often wt
MT64_mt: |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
MT67_wt: |
|
|
|
|
N14 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
MT160_mt: |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
MT101_mt: |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
MT221_wt: |
|
|
|
|
N11 |
|
N28 |
|
|
|
|
|
|
|
MT264_wt: |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
N16 |
|
|
|
|
|
|
|
|
||
MT46_wt: |
|
|
|
|
|
|
N26 |
N31 |
|
|
|
|||
107B_mt: |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
MT60_wt: |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
MT132_mt: |
|
|
|
|
|
MT24_mt: |
|
|
N30 |
N39 |
|
MT54_wt: |
|
|
|
||
|
|
|
|
||
|
|
|
|
|
|
MT21_wt: |
|
N22 |
|
|
|
012.10-NOR: |
|
N18 |
|
|
|
MT5_wt: |
|
N17 |
|
|
|
125.10-NOR: |
|
N9N21 |
|
|
|
020.10-NOR: |
|
|
N29 |
|
|
MT17_wt: |
|
N3 |
|
|
|
MT44_wt: |
|
N25 |
|
|
|
|
N5 N20 |
|
|
||
MT3_mt: |
|
|
|
|
|
MT18_wt: |
|
N15 |
|
N32 |
|
MT19_wt: |
|
N10N7 |
N23 |
|
|
406A_wt: |
|
N6 |
|
|
|
|
|
N49 |
|||
123B_mt: |
|
N4 |
|
||
406B_wt: |
N2 N8 |
|
|
|
|
MT31_wt: |
N1 |
N13N19 |
N27 |
|
|
MT65_mt: |
N0 |
|
|
|
|
|
|
N24 |
|
|
|
011.10-NOR: |
|
|
|
|
|
MT181_mt: |
|
|
|
|
|
017.10-NOR: |
|
|
|
|
|
035.10-NOR: |
|
N12 |
|
|
N55 |
012.20-NOR: |
|
|
|
||
|
|
|
|
||
016.10-NOR: |
|
|
|
|
|
MT38_wt: |
|
|
|
|
|
MT57_wt: |
|
|
|
|
|
MT43_wt: |
|
|
|
N48 |
|
MT20_mt: |
|
|
|
|
|
|
|
|
|
|
|
MT418_mt: |
|
|
|
|
|
MT59_mt: |
|
|
|
N41 |
N53 N58 |
309A_mt: |
|
|
|
||
UT274_mt: |
|
|
|
N33 |
N52 |
MT112_mt: |
|
|
|
|
|
|
|
|
N46 |
|
|
MT161_mt: |
|
|
|
|
|
208A_mt: |
|
|
|
N44 |
|
MT51_wt: |
|
|
|
N42 |
|
UT250_mt: |
|
|
|
|
|
UT065_mt: |
|
|
|
|
N57 |
UT252_mt: |
|
|
|
|
N56 |
101A_mt: |
|
|
|
|
N54 |
UT009_mt: |
|
|
|
|
N51 |
405A_mt: |
|
|
|
N35 |
|
|
|
|
N50 |
||
MT49_wt: |
|
|
|
||
|
|
|
N38 |
||
UT164_mt: |
|
|
|
N47 |
|
MT209_wt: |
|
|
|
|
|
|
|
|
|
|
|
111A_mt: |
|
|
|
N45 |
|
MT29_mt: |
|
|
|
N43 |
|
214A_wt: |
|
|
|
|
|
|
|
|
|
N40 |
|
|
MT61_wt: |
|
|
|
|
|
|
N34 |
N37 |
|
|
|
|
MT342_wt: |
|
|
|
|
|
|
|
N36 |
|
|
|
|
|
|
|
|
|
|
|
|
|||||
111B_mt: |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
sv |
|
p53 |
|
chr8-9 |
|
|
|
|
|
||
p53 status (green = mut, black = wt) Survival (black = low, green = high)
UCSF
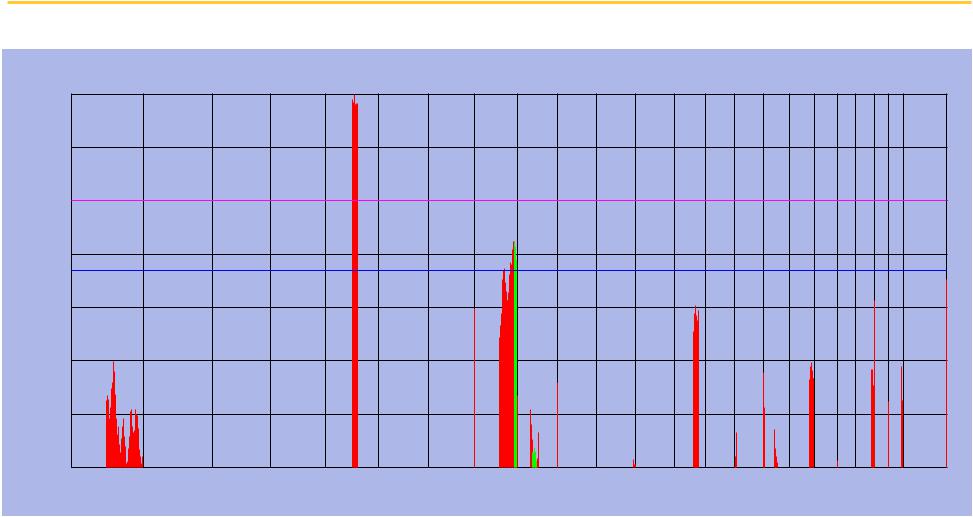
Deletion at 5q11-31 and amplification at 8q24 are correlated with mutant p53
Correlation Magnitude
Correlation magnitude with p53 status
0.35
0.3
0.25
0.2
0.15
0.1
0.05
0
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 20 2122 X |
Genomic Position
Some genes on 5q: APC and IL3

UCSF
Conclusions on permutation and resampling methods
Permutation and resampling methods offer a means to replace complex assumptions with counting.
We can generalize the concept of a statistic to any computable value and apply permutation methods to judge significance.
This can be directly applied in addressing the problem of multiple testing in array-based data.
If we can reduce the number of tests based on an orthogonal observation, we gain statistical power.
Further reading
♦Resampling-Based Multiple Testing : Examples and Methods for P-Value Adjustment by Peter H. Westfall, S. Stanley Young
♦Jain AN, Chin K, Borresen-Dale AL, Erikstein BK, Eynstein Lonning P, Kaaresen R, Gray JW. Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival.Proc Natl Acad Sci U S A. 2001 Jul 3;98(14):7952-7.
♦Dudoit, S., Yang, Y.H., Callow, M.J., and Speed, T. (2000) Statistical methods for identifying differentially expressed genes. Unpublished (Berkeley Stat Dept. Technical Report #578). (To appear: JASA)
♦Tusher V, Tibshirani R, and Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. PNAS 98: 5116-5124.

UCSF
So how do we design array-based experiments?
General scheme
♦Use P samples to screen large number of variables (N) to select a much smaller number (M)
•Expectation, despite multiple comparisons, is that the highest ranked variables contain true effects if they exist
•We must pick M such that for a particular effect size, it is very likely that our M will include a true effect of the specified size
♦On K new samples, screen the M variables in order to identify the true effects with reasonable power
So how do we pick P, N, M, and K?
♦Pick N based on experimental considerations: What pool of variables do you need to consider?
♦Pick P based on practical considerations: you probably won’t be able to pick P large enough to get adequate power.
♦Pick M such that, with preliminary data, the null distribution of the Mth strongest effect makes it very likely that if an effect of the size you want exists, it will be within the top M.
♦Now choose K such that with M variables, you have adequate power to see an effect of the size you want to find.
So how do you choose an effect size?
♦Based on what is of practical significance
♦Note: you can play with the effect size to modulate your power. This is a nasty business though.
