
1Foundation of Mathematical Biology / Foundation of Mathematical Biology
.pdf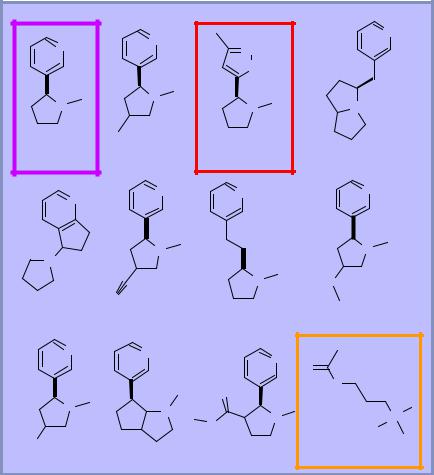
UCSF |
Molecular similarity: 3D |
|
|
|
|
3D similarity
♦Surface-based comparison approach
♦Requires dealing with molecular flexibility and alignment
♦Much slower, but fast enough for practical use
What is the algorithm?
♦Take a sampling of the conformations of molecules A and B
♦For each conformation, optimize the conformation and alignment of the other molecule to maximize S
♦Report the average S for all optimizations
Key issues: not number of atoms. Number of rotatable bonds, alignment
N |
N |
N |
N |
|
|||
|
|
||
|
|
O |
|
N |
N |
N |
N |
|
|||
1.00 |
0.97 |
00..9393 |
00..9191 |
N |
N |
N |
N |
|
|
|
|
N |
N |
|
N |
|
|
|
|
|
N |
N |
O |
|
|
|
0.90 |
0.89 |
|
00..8888 |
00..8787 |
N |
N |
|
N |
O |
|
|
|
||
|
|
|
|
|
|
|
|
O |
O |
N |
N |
|
|
|
O |
N |
N+ |
||
|
|
|
|
HO
0.87 |
0.83 |
00..8282 |
00..6363 |
UCSF |
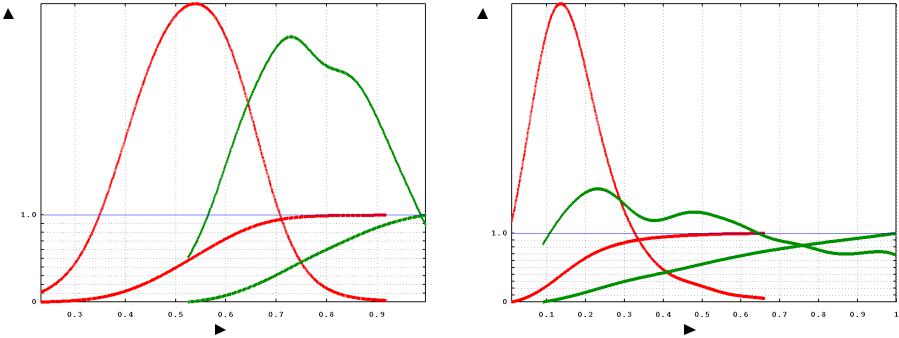
Distributions for the two methods are very different: |
||||
What are the quantitative overlaps? |
|||||
|
|
Molecule pairs observed crystallographically to bind the sameme sitessites |
|||
|
|
|
|
|
|
|
|
Molecule pairs observed crystallographically to bind differentent sitessites |
|||
|
|
|
|
|
|
|
|
|
Morphological Similarity |
|
|
|
Daylight Tanimoto Similarity |
|
|
|
|
|
|||||
|
|
|
|
|
|
|
||
(Probability distribution and integration) |
(Probability distribution and integration) |
|||||||
|
Morphological similarity |
|
|
|
|
Topological similarityy |
|
|
|
|
|||||||
The unrelated pairs distributions are nearly normal
The related pairs distributions are multi-modal, possibly a mixture of normals
UCSF |
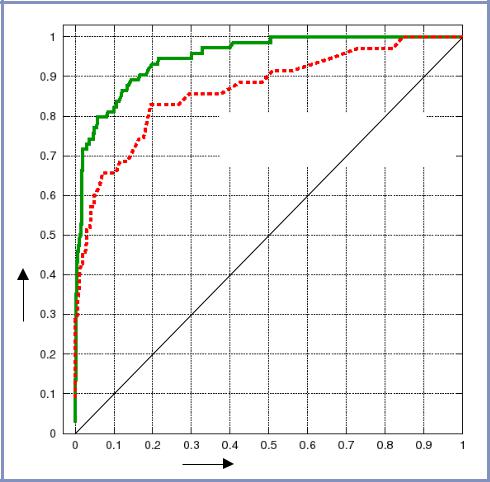
Receiver operator characteristic curves (ROC curves) |
|
plot the relationship of TP rate and FP rate |
||
|
|
|
To construct a ROC curve:
♦Vary the similarity threshold over all observed values
♦At each threshold, compute the proportion of true positives and the proportion of false positives
♦At low thresholds, we should have high FP, but perfect TP
♦At high thresholds, we should have low FP, but poorer TP
At a false positive rate of 0.05, MS yields a 47% reduction in the number of related pairs that are lost
At a true positive rate of 0.70, MS yields a 7-fold better elimination of false positives
Morphological Similarity |
Daylight Tanimoto Similarity |
True positive rate |
False positive rate |
UCSF |
Paired data |
|
|
|
|
Spearman’s rank correlation test
♦We have (Xi,Yi) for n samples
♦We want to know if there is a relationship between the paired samples, but we don’t know if it should be linear, so we need an alternative to Pearson’s r
♦Replace the (Xi,Yi) with (Rank(Xi),Rank(Yi))
♦Compute Pearson’s r for the new values
♦Alternative formulation, where d = difference in ranks for each
data pair |
|
∑d 2 |
|
|
r =1− |
||
|
|
|
|
|
s |
n(n2 |
−1) |
|
|
||

UCSF |
Example: Spearman’s rank correlation |
|
|
|
|
UCSF |
Paired data |
|
|
|
|
Kendall’s Tau: another rank correlation test
♦We have (Xi,Yi) for n samples
♦Definition
•Look at all pairs (Xi,Xj) and corresponding (Yi,Yj)
•Score a 1 for a concordant event
•Score a -1 for a discordant event
•Score 0 for ties in values
•Normalize result based on the number of comparisons
•We get a statistic from -1 to 1
♦Kendall’s Tau has a slightly nicer frequency distribution
♦It can be less sensitive to single outliers

UCSF
Codelet to compute Kendall’s Tau (generalized for real-valued ties)
double k_tau(double *actual, double *predicted, int n, double delta1, double delta2)
{
long int i,j;
double total = 0.0, compare = 0.0;
for (i = 0; i < n; ++i) { for (j = i+1; j < n; ++j) {
compare += 1.0;
/* first check if either is equal --> get no benefit */ if (fabs(actual[i]-actual[j]) <= delta1) {
continue;
}
if (fabs(predicted[i]-predicted[j]) <= delta2) { continue;
}
/* now check if they are correct or incorrect */
if ((actual[i] > actual[j]) && (predicted[i] > predicted[j])) total += 1.0;
else if ((actual[i] < actual[j]) && (predicted[i] < predicted[j])) total += 1.0;
else total += -1.0; /* we have a missed rank match */
}
}
if (compare == 0.0) return(0.0); return(total/compare);
}
UCSF |
Paired data |
|
|
|
|
Signed rank test (Wilcoxon)
♦We have (Xi,Yi) for n samples
♦Definition
•Compute all differences (Xi-Yi)
•Sort them, low to high, based on absolute value
•Assign ranks to each
•Multiple each rank associated with a negative difference by -1
•Sum the negative ranks and positive ranks
•Take the smaller magnitude sum: This is your statistic
♦Again, tables are available for small n
♦An approximation is available for large n
UCSF |
Conclusions: Non-parametric statistics |
|
|
|
|
Non-parametric statistics reduce reliance on distributional assumptions about your data
♦They often give very sensitive tests
♦Generally though, the corresponding parametric tests are more sensitive when it their assumptions hold
♦Note that the process is generally the same
•Compute your statistic
•Look up a significance value or compute one from an approximation
Resampling and permutation-based methods move toward deriving everything from the data observed

UCSF
BP-203: Foundations of Mathematical Biology
Statistics Lecture III: October 30, 2001, 2pm
Instructor: Ajay N. Jain, PhD
Email: ajain@cc.ucsf.edu
Copyright © 2001
All Rights Reserved
