
Cell Biology Protocols
.pdf
PROTOCOL 4.19
Separation of smooth and rough ER in a self-generated iodixanol gradient
Reagents
OptiPrep (60% iodixanol)
Homogenization medium (HM): 0.25 M sucrose, 10 mM Tris-HCl
Light mitochondrial supernatant: Protocol 4.8 (using the above HM) as far as step 8
Include protease inhibitors in the HM as required
Equipment
Ultracentrifuge with 8 × 39 ml fixed-angle (with open-topped thick-walled tubes) and a vertical or near-vertical rotor (e.g. Beckman VTi65.1 or equivalent) and suitable sealed tubes (approx 11 ml) 1
Dounce homogenizer (20–30 ml loosefitting, Wheaton type B)
Gradient collection device for unloading low-density end first 2
Procedure [96]
The method is designed for approx. 10 g
of |
liver. |
Carry out |
all |
procedures at |
|||
0–4 |
◦ |
C. |
3 |
|
|
|
|
|
|
|
|
|
|||
1. |
Centrifuge |
the |
light |
mitochondrial |
|||
|
(15 000g) |
supernatant |
at 100 000g for |
||||
40 min.
2.Discard the supernatant and resuspend
the microsomal pellet in 30 ml of HM using the Dounce homogenizer. 4
3.Mix the microsome suspension with half its volume of OptiPrep (final iodixanol concentration 20%, w/v) and transfer to 11.2 ml Optiseal tubes for the Beckman VTi65.1 vertical rotor.
4. Centrifugation |
at |
350 000gav |
for |
2 h |
at 4 ◦C. Turn |
the |
brake off, |
or |
use |
a slow deceleration program, during below 2000 rpm.
5.Harvest the gradients by upward dis-
placement with a dense medium in 0.5 ml fractions for analysis. 5 6 7
Notes
1 Beckman Optiseal tubes are the most convenient sealed tubes to use and are the recommended ones for this procedure. They are sealed by a central plastic plug that can be removed after centrifugation to allow easy access to the gradient.
2 The tube puncture part of a Beckman Fraction Recovery System or the Labconco Auto Densi-flow is the recommended device for unloading Optiseal tubes. Crimp or heat-sealed tubes are less easy to unload and must be collected dense end first.
3 Although developed for rat liver the method may be used for any tissue.
4 The rough ER in the 100 000g pellet tends to make the latter gelatinous; make sure this is well dispersed.
130 ISOLATION AND FUNCTIONAL ANALYSIS OF ORGANELLES
|
3 |
|
|
|
|
|
|
|
|
50 |
|
|
2.5 |
|
|
|
|
|
|
|
|
40 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
|
|
|
|
|
|
|
|
|
NADPH/min/frac |
|
|
|
|
|
|
|
|
|
|
30 |
|
mg/frac |
1.5 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
20 |
||
|
|
|
|
|
|
|
|
|
|
nmol |
|
|
1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.5 |
|
|
|
|
|
|
|
|
10 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0 |
3 |
5 |
7 |
9 |
11 |
13 |
15 |
17 |
0 |
|
|
1 |
19 |
|
Fraction number
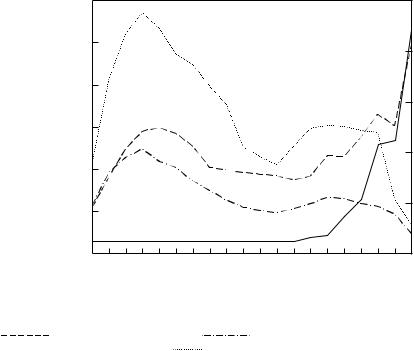
Figure 4.2 |
Fractionation of rat liver microsomes in a self-generated |
iodixanol gradient. Starting |
||||
concentration of iodixanol = 20%; |
Beckman |
VTi65.1 vertical rotor at |
350 000g for 2 h. Protein |
|||
( |
) mg/fraction, phospholipids ( |
) mg/fraction, RNA ( |
|
|
) mg/fraction (×5), |
|
|
|
|||||
NADPH cytochrome c reductase ( |
) |
|
|
|
|
|
5 The bottom of the gradient is extremely viscous, due to the high concentration of iodixanol and the presence of RNA, so collection of the gradient dense end first is not really the best option.
6 For more information on the harvesting and analysis of gradients, see ref. 18.
7 A typical profile of major markers for the SER and RER is given in Figure 4.2. Protein, lipid and NADH
cytochrome c reductase profiles of the gradient reveal two broad bands of material. RNA increases gradually in the lower third of the gradient, the very sharp increase in the bottom fraction being accompanied by a rapid fall in phospholipid and NADH cytochrome c reductase. The SER is thus distributed broadly in the top half of the gradient, the RER in the bottom third and ribosomes band in the last fraction.

PROTOCOL 4.20
NADPH-cytochrome c reductase assay [50]
Reagents
Buffer: 50 mM phosphate, pH 7.7, containing 0.1 mM EDTA
Cytochrome c (25 mg/ml) in buffer (make up fresh; keep on ice)
NADPH (2 mg/ml) in buffer (make up fresh; keep away from light; keep on ice)
Rotenone: 1 mg/ml in ethanol
of buffer, then mix well with up to
50 µl of sample.
3.When the absorbance at 550 nm is steady, add 0.1 ml of NADPH.
4.Mix well and continue to record the absorbance until a linear increase in
value can be measured over a period of 1–2 min. 2
Equipment
Recording spectrophotometer (visible wavelength) with 1 ml cuvettes. 1
Procedure
The procedure is given for a single-beam spectrophotometer linked up to a chart recorder (0.2 absorbance units full-scale deflection).
1.Bring the buffer to room temperature and carry out all operations at this temperature.
2.In a 1 ml cuvette add 50 µl of cytochrome c and 10 µl of rotenone to 1 ml
Notes
1 A dual-beam spectrophotometer is perhaps the best instrument; if one is available, replace the NADPH with buffer in the blank cuvette. Single beam instruments linked up to a chart recorder (used in this procedure) will probably be the most widely available. Even non-recording spectrophotometers can be used, although the measurement is rather more tedious.
2 Calculate the enzyme activity in terms of µmoles cytochrome c reduced; the molar extinction coefficient of reduced cytochrome c is 27 000.

PROTOCOL 4.21
Glucose-6-phosphatase assay [49]
Reagents
2.5% (w/v) Ammonium molybdate in
2.5 M H2SO4
Fiske-Subbarow reducing solution (prepare and store as per manufacturer’s instructions)
Substrate solution: 0.1 M glucose-6-phos- phate
Buffer: 5 mM EDTA, 20 mM histidineHCl buffer pH 6.5
8% (w/v) Trichloroacetic acid (TCA), keep ice-cold
Equipment
Centrifuge (low-speed refrigerated) with swinging-bucket rotor to hold the assay tubes
Spectrophotometer (visible wavelength) Tubes, approx. 4 ml 1
Water baths set at 37 and 100 ◦C
Procedure
1.Prepare the test solution: mix substrate and buffer solution in the ratio 2.5 : 2 (v/v).
2.Add 50 µl of sample to 0.45 ml of the test solution and mix. For each sample also set up a control tube containing only 0.45 ml of test solution.
3.Incubate the tubes at 37 ◦C for 30 min.
4.Transfer the tubes to an ice/water bath;
add 50 µl of sample to the control tubes.
5.Add 2.5 ml of ice-cold TCA to all tubes.
6.Keep at 0–4 ◦C for 20 min. 2
7.Centrifuge all the TCA containing tubes at 1000g for 15 min at 4 ◦C.
8.Remove 1 ml of supernatant to a new tube.
9.Add 1.15 ml water, 0.25 ml of ammonium molybdate solution and 0.1 ml of reducing solution.
10.Heat all tubes at 100 ◦C for 10 min.
11.Cool and read absorbance of all test solutions at 820 nm against the appropriate controls.
Notes
This procedure takes 2–3 h.
1 Enzyme assays based on the measurement of released phosphate can be a problem if care is not taken to eliminate, as far as possible, phosphate contamination from the tubes and bottles used for the reagents. Glass tubes and glass containers must be acid-washed and then rinsed three times in distilled water before use. Plastic tubes used for the incubation at 37 ◦C need to be checked for absence of phosphate.
2 Longer times are not harmful.

PROTOCOL 4.22
RNA analysis [97]
Reagents
Orcinol reagent: 0.5 g orcinol, 0.25 g FeCl3.6H2O in 50 ml concentrated HCl (make up freshly and keep at 4 ◦C until required)
Trichloroacetic acid (TCA) solution: 20% (w/v) in water
RNA standard: 250 µg/ml in water
Equipment
Microcentrifuge and 1.5–2.0 ml microcentrifuge tubes
Water bath
Spectrophotometer
Procedure
1.Dilute RNA stock to give 50, 100, 150 and 200 µg/ml solutions.
2.Transfer 0.5 ml of each RNA standard (50–250 µg/ml) and a water blank to microcentrifuge tubes.
3.Dilute gradient samples in microcentrifuge tubes to 0.5 ml with water.
4.Add TCA to all tubes so that the final concentration is 5%.
5. Heat in a water bath at 90 ◦C for
20 min.
6.Microcentrifuge for 2 min.
7.Transfer the supernatant to another tube and add an equal volume of the orcinol reagent.
8.After 20 min at 100 ◦C, cool and measure the absorbance at 660 nm. 1
Note
The procedure will take approx. 1.5 h.
1 Neither Nycodenz nor iodixanol interfere with this assay.

PROTOCOL 4.23
Isolation of Golgi membranes from liver [52]
Reagents
Homogenization buffer (HM): 0.5 M sucrose, 1% (w/v) dextran (Mr = 225 000) in 0.05 M Tris-maleate, pH 6.4
Density barrier: 1.2 M sucrose, 0.05 M Tris-maleate, pH 6.4
Include protease inhibitors in these two solutions as required
Equipment
High-speed centrifuge with swingingbucket rotor for 30–50 ml clear plastic tubes. 1
Phase contrast microscope
Polytron homogenizer
Syringe |
(10 or 20 ml) and metal can- |
nula |
2 |
Ultracentrifuge with swinging-bucket rotor for 17 ml tubes
Glass rod
3.Centrifuge for 15 min at 5000g and
very carefully aspirate the supernatant using the syringe and cannula. 4
4.The Golgi membranes are contained in the upper (yellow-brown) portion of the bipartite pellet; resuspend them in some of the residual supernatant by very gentle stirring with a glass rod. Be very careful to avoid resuspension of the lower part of the pellet, which contains nuclei, some heavy mitochondria and whole cells.
5.Transfer the resuspended Golgi material to a new tube using the syringe and cannula.
6.Adjust the concentration of the suspension with HM to 6 ml per 10 g liver and layer over 2 volumes of 1.2 M sucrose in a tube for the swinging-bucket rotor.
7.Centrifuge at 120 000g for 30 min.
8.Remove the Golgi membranes that
collect at the interface with a syringe and cannula. 5
Procedure
Carry out all operations at 0–4 ◦C. The procedure is for a single rat liver (approx. 10 g).
1.Mince the liver finely using a razor blade and suspend in HM (about 10 g liver per 20 ml buffer).
2.Homogenize in a Polytron homoge-
nizer set at 10 000 rpm (setting 1) for 30–50 s. 3
Notes
This procedure will take approx. 1.5 h.
1 A fixed-angle rotor is not an option because of the difficulty such a rotor would pose for step 4.
2 Metal ‘filling’ cannulas can be obtained from any surgical equipment supplies company.
3 Use the Polytron in 10 s ‘bursts’, interposed by 20 s ‘rests’ to avoid any heating. Monitor the progress of the homogenization by phase contrast microscopy.
4 Keep the tip of the cannula close to the meniscus.
5 The dextran in the HM maintains the stacking of the Golgi tubules. If it is necessary to isolate the various domains from the Golgi, the stacks
PROTOCOL 4.23 |
135 |
must be disaggregated by |
hydroly |
sis of the dextran. This is achieved by incubating the isolated Golgi in 4–5 ml of HM with 3 mg each of α- amylase Type X-A from Aspergillus oryzae and α-amylase Type III-A from barley for 45 min at 4 ◦C. Destacking is completed by gentle liquid shearing through a Pasteur pipette (tip i.d. = 1 mm) or through a syringe with metal cannula [98].

PROTOCOL 4.24
Assay of UDP-galactose galactosyl transferase [54]
Reagents
Keep all solutions on ice.
Acceptor solution: 10 mM ATP, 5% (w/v) ovalbumin in assay buffer (make up fresh)
Assay buffer: 10 mM MnCl2, 30 mM 2-mercaptoethanol, 0.1 M cacodylate buffer, pH 6.2, containing 0.2% (w/v) Triton-X100
TCA (10%, w/v)
UDP-gal: |
12.5 mM UDP-galactose |
containing |
UDP-[6-3H]-galactose |
(70 kBq/ml) Scintillant 1
2.Pin the discs to a polystyrene board, making sure that the discs do not touch the board.
3.Place 250 ml of 10% TCA in a beaker in ice.
4.In 0.3 ml assay tubes (in an ice-water bath), mix 50 µl of acceptor solution with 50 µl sample and add 10 µl radiolabelled UDP-gal solution.
5.At zero time and after a 20 min incubation at 37 ◦C transfer 50 µl of the incubation mixture to a filter disc.
6.Plunge the discs into the ice-cold TCA. 2
7.Leave for at least 2 h, swirling the discs occasionally.
Equipment
Filter paper discs (2.4 cm)
Ice-water bath
Plastic assay tubes (0.3 ml)
Polystyrene board with pins
Scintillation counter
Water bath at 37 ◦C
Procedure
1. Prepare two filter paper discs (2.4 cm) per sample (numbered lightly with a pencil).
8.Wash the discs in several changes of distilled water; leave to dry overnight at room temperature.
9.Count in a suitable scintillant.
Notes
This procedure will take 18–20 h.
1 Any commercial scintillant for nonaqueous samples will suffice.
2 Do not allow the discs to float on the TCA; the pencil mark is liable to come off.

PROTOCOL 4.25
Purification of human erythrocyte ‘ghosts’ [55]
Reagents
Human blood (0.38% citrate or 1 mM EDTA as anticoagulant)
Hypotonic buffer: dilute isotonic buffer 15.5× with distilled water
Isotonic buffer: mix 0.155 M NaH2PO4 and 0.103 M Na2HPO4 to pH 7.4
Equipment
High-speed centrifuge with fixed-angle rotor (8 × 50 ml)
Low-speed (refrigerated) centrifuge with swinging-bucket rotor (50 ml tubes)
Wide-bore pipette attached to automatic pipette-filler
Procedure
Carry out all operations at 0–4 ◦C. The procedure is for 30–40 ml of blood.
1.Centrifuge the blood in a capped tube at 800g for 15 min. 1
2.Aspirate the plasma and buffy coat into a trap containing disinfectant.
3.Add isotonic buffer to the original volume and gently resuspend the erythrocyte pellet using the wide-bore pipette.
4.Centrifuge the erythrocyte suspension at 800g for 15 min.
5.Repeat steps 2–4 three more times. 2
6. After removing the |
buffer |
from |
|
the final |
wash, resuspend the ery- |
||
throcytes |
in isotonic |
buffer |
(32 ml |
final volume).
7.Place 28 ml of hypotonic buffer in each of 16 tubes for the high-speed centrifuge and from an automatic
pipette quickly add 2 ml of the washed erythrocyte suspension. 3
8.Centrifuge at 20 000g for 20 min.
9.Aspirate the supernatant and resuspend the erythrocyte ‘ghost’ pellet in the
hypotonic buffer by gentle pipetting, avoiding any hard-packed buttons. 4
10.Repeat the centrifugation and washing procedure three times.
11.Finally, resuspend the washed ‘ghosts’ in a suitable buffer.
Notes
The procedure will take approx. 3 h.
1 Operators need to be properly trained in the handling of human blood.
2 Significant numbers of leukocytes in the final erythrocyte suspension can cause serious aggregation due to lysis of their nuclei during the hypotonic buffer steps. It is therefore important that as much of the leukocyte fraction, which forms the buffy coat, is removed, even at the expense of losing some of the erythrocytes.
138 ISOLATION AND FUNCTIONAL ANALYSIS OF ORGANELLES
3 Effective lysis of the erythrocytes only occurs if the cells are rapidly exposed to the hypotonic buffer.
4 The hard-packed button contains aggregated material (debris, partially
disrupted leukocytes and leukocyte nuclei). This must be avoided in the resuspension of the erythrocyte ghosts.
