
Solid-Phase Synthesis and Combinatorial Technologies
.pdf
2.1 PEPTIDES 53
L2
L |
7 |
|
L3 |
Tyr |
L |
|
1 |
Asp 8 |
4 |
||||
|
Glu |
|
Asn |
|
Glu 10 |
|
|
6 |
|
|
|
|
|
|
Pro |
|
Arg |
|
Ala |
|
|
|
D-Phe |
Ala |
|
|
|
|
|
2.12 |
|
|
|
|
best results obtained with L3: PAL linker, 71% final yield
Figure 2.5 Structure of the cyclic peptide 2.12 and its possible side-chain attachments onto the support.
This synthesis has been recently revisited (5), and a new linker that allows the fastening of a backbone amide onto the support strategy has been applied. While this peptide represents a nice target that allows direct comparison of the quality of the SPS with previous attempts, its main advantage lies in the fact that use of the backbone amide linkage, or BAL (2.13, Fig. 2.6), allows anchoring of peptides other than Asp, Glu, Gln, or Asn, thus permitting the synthesis of analogues otherwise not accessible in the original work.
The linker was prepared by hooking the aldehyde 2.14 (68) onto the support and then performing a reductive amination with the first α-amino acid protected as an allyl
|
|
O |
|
OMe |
|
H |
|
|
|
|
|
|
|
|
|
|
O |
OMe |
|
|
|
NH2 + |
|
|
a |
N |
|
||
|
|
|
|
O |
|
|
+ |
||
|
|
|
|
|
|
|
|
||
|
|
OMe |
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
2.14 |
COOH |
|
|
OMe O |
|
|
|
|
|
|
|
|
|||
|
|
Me |
|
|
H |
|
|
|
|
|
|
|
|
|
O |
|
OMe |
|
|
|
+ |
OAllyl |
b |
|
N |
|
O |
||
|
|
|
|
|
|
||||
+ |
H3N |
|
|
|
|
O |
|
H |
|
Cl- |
O |
|
|
|
|
|
|||
|
|
|
|
|
N |
OAllyl |
|||
|
|
|
|
|
2.13 |
|
|
|
|
|
|
|
|
|
L-Ala-BAL linker |
OMe |
|
Me |
|
|
|
|
|
|
|
|
|
||
a) HATU/DIEA, DMF; b) NaBH3CN, DMF.
Figure 2.6 Synthesis of the BAL linker 2.13.

54 SOLID-PHASE SYNTHESIS: OLIGOMERIC MOLECULES
ester (Ala in this example). The peptide chain was grown in the C-to-N direction using Fmoc-protected amino acids. Acid-labile side-chain protection was used for Arg (Pmc), Glu and Asp (OtBu), Asn (Trt), and Tyr (tBu).
Attempts to incorporate the second amino acid were frustrated by the formation of the diketopiperazine 2.15 (Fig 2.7). Apparently, during the standard Fmoc protocol,
H |
O |
OMe |
N |
||
O |
|
H |
|
N |
|
|
2.13 |
OMe |
|
|
H
N
O
H
N
O
H
N
O
2.15
|
O |
|
|
Me |
|
OAllyl |
+ Fmoc |
||
|
|
N COOH |
||
Me |
|
|
|
H |
|
|
|
|
|
|
a |
|
|
Fmoc |
|
|
|
|
|
|
|
|
|
NH |
O |
|
OMe O |
Me |
|
|
|
|
||
|
|
|
|
|
|
|
|
N |
Me |
|
OMe |
|
O |
OAllyl |
|
|
|
||
|
b |
|
|
|
|
|
|
|
NH2 |
O |
|
|
OMe O |
Me |
|
|
|
||
|
|
|
|
|
|
|
|
N |
Me |
|
OMe |
|
O |
OAllyl |
|
|
|
||
|
O |
O |
OMe |
Me |
|
|
NH |
N
Me
OMe O
a) HATU/DIEA, DMF; b) 20% piperidine in DMF.
Figure 2.7 Side reaction of BAL-supported N-Fmoc amino acids during Fmoc deprotection: Diketopiperazines 2.15.

H |
O |
OMe |
N |
||
|
|
O |
O |
|
H |
|
N |
2.13
OMe |
Me |
H |
O |
N |
|
O |
|
2.16 |
|
H |
O |
N |
|
|
O |
2.1 PEPTIDES 55
|
|
Me |
OAllyl |
+ |
Tt |
|
N COOH |
|
|
|
H |
a,b
OMe
NH3+
O
Me
N Me
OMe
O OAllyl
c |
O |
||
|
|
|
H |
|
|
|
N |
|
|
|
|
NH Fmoc
OMe
O
Me
N Me COOtBu
OMe
O OAllyl
d,c (7 cycles)
|
|
H |
O |
|
OMe |
|
|
|
|
|
|
|
|
|
|
|||||
|
|
N |
|
|
|
OAllyl OtBu |
Tt |
OtBu |
|
|
|
|||||||||
|
|
|
|
|
|
|
|
|
|
|||||||||||
|
|
O |
|
|
|
|
|
|
Ala |
|
|
|
Asn |
|
|
|
|
|
||
|
|
|
|
Glu |
|
Glu |
D-Phe |
|||||||||||||
|
|
|
|
|
|
|
|
|
Ala |
Tyr Asp Pro Arg Fmoc |
||||||||||
|
|
e |
|
OMe |
|
|
|
|
|
|
OtBu |
|
OtBu |
|
|
Pmc |
||||
|
|
|
|
|
|
|
|
|
|
|
||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
f,d,g |
|
|
|
|
|
|
|
|
|
|
|||
|
|
D-PheGlu Asn Glu Ala |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Fmoc |
|
Arg Pro Asp Tyr Ala |
OAllyl |
OtBu Tt |
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
||||||||||
2.17 |
|
|
|
|
|
|
|
|
|
|
|
|||||||||
|
|
|
OtBu |
Asn |
|
|
OtBu |
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
Glu |
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
Ala |
|
Tyr |
Asp |
|
|
|
|
|
|
|
|
||||||
|
|
|
BAL |
Ala |
D-Phe |
Glu |
|
|
|
|
|
|
|
|
||||||
|
|
|
Arg |
|
|
|
Pro |
|
|
OtBu |
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
Resin-bound, protected 2.12 |
|||||||||||
|
|
|
|
Pmc |
|
|
|
|
|
|
|
|
|
|
||||||
a:PyAOP, DIEA, DMF, rt, 2 hrs; b: TFA/H2O/DCM 2/1/97, rt, 5 ';
c:Fmoc-AA-OH, PyAOP, DIEA, DMF, rt, 2 hrs; d: 20% piperidine, DMF;
e:TFA/Et3SiH/H2O 92/5/3, rt, 2 hrs; f: Pd(PPh3)4, DMSO/THF/0.5N HCl/morpholine 2/2/1/0.1, rt, 3 hrs;
g:PyAOP, HOAt, DIEA, DCM, rt, 2 hrs.
Figure 2.8 SPS of resin-bound fully protected cyclo(Arg-D-Phe-Pro-Glu-Asp-Asn-Tyr-Glu- Ala-Ala). 2.12 and release of deprotected 2.17.

56 SOLID-PHASE SYNTHESIS: OLIGOMERIC MOLECULES
the deprotection gave a free nucleophilic amine that intramolecularly attacked the allyl ester to produce the unwanted diketopiperazine 2.15. Although the use of a t-butyl ester prevented diketopiperazine formation, the need for a protection for the C-terminus that was removable under mild conditions and was orthogonal to the protected acid side chains did not suggest this selection.
It was found that the trityl (Tt) group was suitable for the amine in that it could be removed selectively after coupling to give a protonated amine 2.16 that was incapable of cyclization (Fig. 2.8). The SPS continued with standard Fmoc-based coupling/deprotection cycles using PyAOP [phosphonium tri-1-pyrrolidinyl (3H-1,2,3-tria- zolo[4,5-b] pyridin-3-yloxy)-hexafluoro phosphate] (69) [a phosphonium salt derived from HOAt (hydroxy benzo triazole)] as coupling agent together with DIEA to improve the reaction yields (8). A sample of the open decapeptide 2.17 was cleaved in greater than 85% and characterized off-bead as the N-Fmoc C-allyl ester derivative.
The cyclization on SP under a variety of experimental conditions was evaluated by monitoring the amounts of noncyclized residual decapeptide, the total yield of the desired product, and the degree of C-terminal epimerization due to the resin-bound activation of the carboxylic group. The results are summarized in Table 2.1. The best results were obtained using DIEA as base and DCM as solvent while other combinations of solvents and bases produced more extensive racemization (entries b, c, d, e) or significant amounts of open decapeptide (entries c, e).
The synthesis of the cyclic peptide 2.12 on SP was achieved in excellent yield and purity, demonstrating the flexibility of the SPS approach for a complex sequence containing difficult amino acids such as Glu, Asp, and Arg. A new linker strategy suitable for any peptidelike sequence with no problems of stability and able to permit the final modification of the C-terminus was developed. The iterative nature of peptide SPS and the robustness of these oligomeric structures were instrumental in reaching such good-quality results that are easily amenable to automated SPS procedures.
TABLE 2.1 Cyclization of the Linear Decapeptide on SP: Optimization Studies
Cyclization |
Base |
Solvent |
Noncyclized |
L–D |
Conditionsa |
Peptideb |
ratio/epimerization |
||
|
|
|
|
|
a |
DIEA |
DCM |
ND |
88/12 |
b |
DIEA |
DMF |
ND |
75/25 |
c |
2,4,6-triMepyridine |
DCM |
26% |
76/24 |
d |
2,4,6-triMepyridine |
DMF |
ND |
50/50 |
e |
2,4,6-triMepyridine |
CHCl3/TFE 7/3 |
75 |
78/22 |
aPyAOP–HOAt as coupling agents. bND = not detectable.
2.2 OLIGONUCLEOTIDES 57
2.2 OLIGONUCLEOTIDES
2.2.1 Solid-Phase Synthesis of Oligonucleotides
Oligonucleotides (ONs) are the other major class of important biopolymers that readily lends itself to automated synthesis on SP and as such has also received a great deal of attention, the first attempts (70) dating back to the mid 1960s. While the SPS of this class of natural oligomers has the same general advantages seen for peptides, the different properties of the two oligomeric backbones lead to different choices in terms of supports, linkers, protecting group strategy, and cleavage conditions.
There are three widely used chemistries for oligodeoxynucleotide synthesis, and all have been successfully transferred onto SP The first is the so-called phosphotriester approach (71), which is shown in Fig. 2.9. In this method a 5′-protected nucleoside
HO |
|
P1O |
P1O |
|
O |
NB(P3) |
O NB(P ) |
O |
NB(P3) |
|
|
|||
|
|
3 |
|
|
L |
3' |
|
|
+ |
|
|
|
a |
|
O |
|
|
|
O |
|
|
O |
|
|
|
|
OP2 |
|
||||
|
|
|
|
|
P |
|
|
|
|
P |
|
||
|
2.18 |
|
|
O |
OP2 |
2.19 |
|
|
O |
|
O |
NB(P3) |
|
|
|
|
O |
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
b,a |
|
|
L |
O |
3' |
|
|
P1O |
|
|
|
|
|
(n-1 cycles) |
|
|
|
|
|
|
|
O |
NB(P3) |
|
|
|
|
|
|
|
2.20 |
|||
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
O O |
|
|
|
|
|
|
|
|
|
|
|
|
|
P |
OP2 |
|
|
|
|
|
|
|
|
|
|
|
|
O |
|
|
|
|
HO |
|
|
|
|
|
||
|
O |
|
|
|
|
|
|
|
|
|
|
||
|
|
|
NB(P3) |
|
|
|
|
O |
|
|
|
||
|
|
|
|
|
|
|
|
|
NB |
|
|||
|
|
|
|
|
|
|
|
|
|
|
|||
|
O |
O |
|
|
n-2 |
|
c,d,e |
O |
|
|
|
|
|
|
|
|
|
|
|
O |
|
|
|
||||
|
P |
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
P |
|
|
|
|
|||
|
O |
OP2 |
|
|
|
|
OH |
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
O |
NB(P3) |
|
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
O |
|
NB |
|
||
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
L |
O |
|
3' |
|
|
|
|
O O |
|
|
||
|
|
|
2.21 |
|
|
|
|
|
n-2 |
||||
|
|
|
|
|
|
|
P |
|
|
|
|||
|
|
|
|
|
|
|
|
O |
OH |
|
|
||
|
|
|
|
|
|
|
|
|
|
O |
NB |
||
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
HO |
|
3' |
|
L = LINKER
a:sulfonyl chloride, 1-methylimidazole (cat); b: P1 deprotection; c: P2 deprotection;
d:P3 deprotection; e: cleavage from the support.
Figure 2.9 Phosphotriester protocol for oligonucleotide SPS.
58 SOLID-PHASE SYNTHESIS: OLIGOMERIC MOLECULES
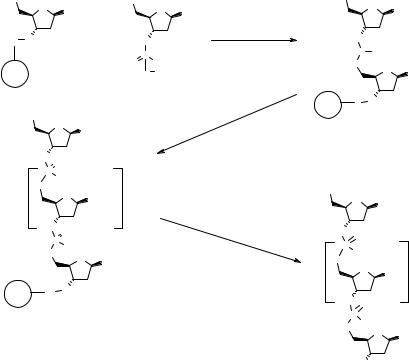
phosphate diester 2.19 is reacted with a 3′-resin-bound nucleoside 2.18 in the presence of an appropriate sulfonyl chloride and 1-methyl imidazole as a catalyst to give the dinucleoside phosphate triester 2.20. Deprotection of the 5′-protecting group gives the bound 5′-OH substrate ready for the next coupling reaction, and so on, until the fully protected target resin-bound oligonucleotide 2.21 is obtained. The phosphate ester protecting group (P2, Fig. 2.9) is hydrolyzed in basic conditions, the protecting groups on the bases are removed, and finally, the nucleotide is cleaved from the resin. This method, which is still used sometimes for specific applications in SPS (72), has been superceded by the phosphonate and the phosphoramidite routes (see below).
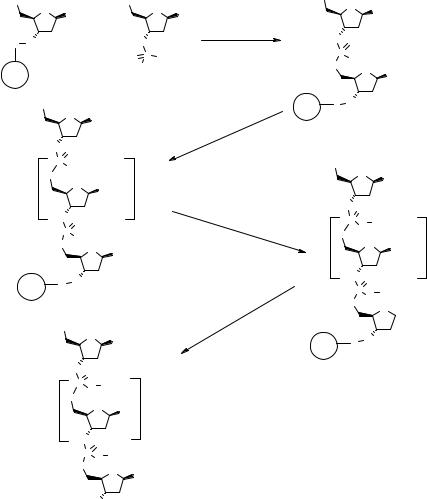
The phosphonate route (73), which is shown in Fig. 2.10, involves the coupling of a 5′-protected nucleoside hydrogen phosphonate 2.22 with the 3′-resin-bound nucleoside 2.18 in the presence of adamantoyl chloride as activating agent to give the 5′-protected dinucleoside phosphonate 2.23. The cycle continues with 5′-deprotection followed by coupling with another nucleoside (n – 1 cycles). The final phosphonate is oxidized with iodine in a basic environment to give the fully protected target ON 2.24. The free oligonucleotide is obtained after deprotection of the nucleotide bases and cleavage from the solid support. This protocol has been successfully automated on SP (74), but the oxidation step is sometimes problematic and the total yields are generally lower than using the phosphoramidite routes, thus confining this method to short oligonucleotide sequences.
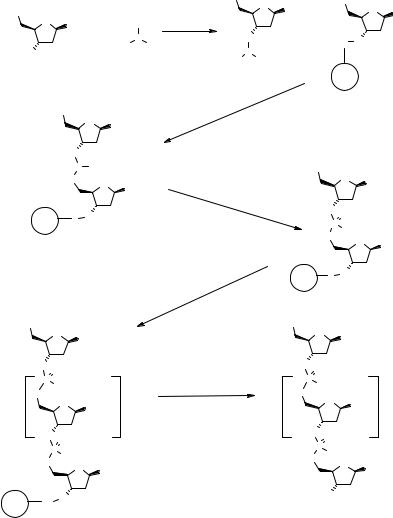
While the SPS phosphonate route is used for specific applications (75, 76), the phosphoramidite method (Fig. 2.11) is the most used method for the assembly of oligonucleotides both in solution (77) and on SP (78). The 5′-protected nucleoside phosphoramidite 2.25, which is readily prepared from a 5′-protected 3′-hydroxy nucleoside and an amino chloromethoxyphosphine, is coupled with the resin-bound nucleoside 2.18 using tetrazole as an activating agent. The resulting phosphite is oxidized with t-butyl peroxide to produce the phosphite triester 2.26. This compound is iteratively extended by deprotection/coupling/oxidation cycles to produce a fully protected ON 2.27 that is subjected to final deprotections before cleavage from the support. This method has been reviewed recently (79–81) and will now be considered in depth.
Many solid supports including macroporous resins (82), cellulose (83), and hydrophobic (84) and hybrid hydrophilic PS resins (72) have been used for the preparation of ONs. Early experiments (85) highlighted the incompatibility of hydrophobic supports with the chemistry used, and among the hydrophilic supports examined the fully inorganic silica-based CPG supports (78) rapidly became the most popular. They are nonswelling materials (see Section 1.1.3) and are ideally suited to continuous-flow SPS. A large variety of pre-packed CPG columns are now commercially available and are regularly employed for the automated synthesis of ONs with more than 100 residues. Over time, the optimization of the CPG structure and of the linker between the nucleoside and the silica core (86–88) has produced supports with pore sizes of 500–1000 Å and fully extended spacers of considerable lengths (>25 atoms). A typical example is the so-called long-chain alkylamine (LCAA) CPG support 2.28 (89) shown in Fig. 2.12. A recent contribution has introduced a novel polymer colloid supported
HO |
|
|
P1O |
|
O |
|
O |
NB(P2) |
|
|
NB(P2) |
||
|
|
|
|
|||
|
|
+ |
|
|
|
a |
L O 3' |
|
|
|
|
O |
|
|
|
|
O |
P OH |
|
|
|
|
|
|
H |
|
|
2.18 |
|
|
|
2.22 |
||
P1O |
O |
|
|
|
|
b,a |
|
NB(P2) |
|
|
(n-1 cycles) |
||
|
|
|
|
|||
O |
O |
|
|
|
|
|
P |
H |
|
|
|
|
|
O |
|
|
|
|
|
|
O |
|
|
|
|
|
|
|
NB(P2) |
|
|
|
||
|
|
|
|
|
||
O |
O |
|
|
|
n-2 |
c |
|
|
|
|
|||
P |
|
|
|
|
|
|
O |
H |
|
|
|
|
|
|
O |
NB(P2) |
|
|||
|
|
|
||||
|
|
|
|
|||
L |
O |
3' |
|
|
|
|
|
|
|
|
|
|
d,e |
HO |
O |
|
|
|
|
|
|
|
NB |
|
|
|
|
|
|
|
|
|
|
|
|
O |
O |
|
|
|
|
|
|
|
|
|
|
|
|
P |
O |
|
|
|
|
|
O |
|
|
|
|
|
|
O |
|
|
|
|
|
|
|
NB |
|
|
|
|
|
|
|
|
|
|
|
|
O |
O |
|
|
n-2 |
|
|
P |
|
|
|
|
|
|
O |
O |
|
|
|
|
|
|
O |
NB |
|
||
|
|
|
|
|||
|
|
|
|
|
||
|
|
HO |
3' |
|
|
|
L = LINKER
2.2 OLIGONUCLEOTIDES 59
P1O
|
O |
NB(P2) |
|
|
|
|
|
||
O |
O |
|
|
|
P |
H |
|
|
|
O |
O |
|
|
|
|
NB(P2) |
|
||
|
|
|
||
|
|
|
|
|
L |
O |
3' |
|
|
|
|
2.23 |
|
|
P1O |
|
O |
|
|
|
|
NB(P2) |
|
|
|
|
|
|
|
|
O |
O |
|
|
|
P |
O |
|
|
O |
|
|
||
O |
|
|
||
|
|
NB(P2) |
|
|
|
|
|
|
|
|
O |
O |
|
n-2 |
|
P |
|
|
|
O
O
O  NB(P2)
NB(P2)
L O 3'
2.24
a:adamantoyl chloride; b: P1 deprotection; c: I2 / pyridine/H2O; d: P2 deprotection;
e:cleavage from the support.
Figure 2.10 Phosphonate protocol for oligonucleotide SPS.
onto a polyethylene filter disk as an alternative support to CPG and resin beads in ON SPS with at least similar synthetic performances (90).
In contrast to SP peptide synthesis, the low acid stability of the ON backbone requires a different linker and chemistry strategy and the growing ON is attached to the support via base-labile bonds. Some examples of these linkers are shown in Fig.
60 SOLID-PHASE SYNTHESIS: OLIGOMERIC MOLECULES
|
|
|
|
|
|
|
|
P1O |
O |
NB(P3) |
HO |
|
O NB(P |
|
|
P |
O |
|
|
|
|
|
|
|
|
|
|
) |
|||
|
|
|
|
|
|
|
|
|
|
|
|||||
1 |
O |
|
NB(P3) |
|
|
Cl |
a |
|
|
+ |
|
|
3 |
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
+ |
|
P2O |
P |
NR2 |
O |
|
|
L |
O |
3' |
|
|
|
|
|
|
|
|
P |
|
|
|
|
||||
|
HO |
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
R2N |
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
OP2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2.25 |
|
|
|
|
2.18 |
|
|
P1O |
|
|
|
|
|
b |
|
|
|
|
|
|
|
|
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
NB(P3) |
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
P |
OP2 |
|
|
|
|
|
P1O |
O |
|
|
|
|
|
|
|
O |
O |
NB(P3) |
|
|
|
|
NB(P3) |
|
||||
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
c |
|
O O |
|
|
|
|
|
|
|
L |
O 3' |
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
P |
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
OP2 |
|
|
||
|
|
|
|
|
|
|
|
|
|
|
O |
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|
|
O |
NB(P3) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
d,b,c |
|
|
L |
O |
3' |
|
|
|
|
|
|
|
|
|
|
(n-1 cycles) |
|
|
|
|
|
2.26 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
P1O |
O |
|
|
|
|
|
|
|
HO |
O |
|
|
|
|
|
|
NB(P3) |
|
|
|
|
|
|
NB |
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
O O |
|
|
|
|
|
|
|
O O |
|
|
|
|
||
|
P |
OP2 |
|
|
|
|
d,e,f,g |
|
P |
OH |
|
|
|
|
|
|
O |
|
|
|
|
|
|
O |
|
|
|
|
|||
|
O |
|
|
|
|
|
|
O |
|
NB |
|
|
|||
|
|
NB(P3) |
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
O |
O |
|
|
|
n-2 |
|
|
O O |
|
|
n-2 |
|
||
|
P |
|
|
|
|
|
|
|
P |
|
|
|
|
||
|
|
|
|
|
|
|
|
|
OH |
|
|
|
|
||
|
O |
OP2 |
|
|
|
|
|
|
O |
|
|
|
|
||
|
|
|
|
|
|
|
|
O |
|
|
|
||||
|
|
O |
NB(P3) |
|
|
|
|
|
|
NB |
|
||||
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
L |
O |
|
3' |
2.27 |
|
|
|
|
HO 3' |
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
L = LINKER
a:DIEA, DCM; b: 1H-tetrazole, MeCN; c: oxidation; d: P1 deprotection; e: P2 deprotection;
f:P3 deprotection; g: cleavage from the support.
Figure 2.11 Phosphoramidite protocol for oligonucleotide SPS.
2.13. An ester bond is widely used, and the succinate 2.29 (91) is one of the most popular linkers. It is introduced onto the LCAA-CPG support as the 3′-monosuccinate ester of the first nucleoside, and it is cleaved under basic conditions in the final SPS step. The inclusion of a sarcosine spacer as in 2.30 (92) increases the linker stability

|
|
|
2.2 OLIGONUCLEOTIDES 61 |
OMe |
|
|
O |
CPG Si |
O |
O |
NH2 |
OMe |
|
|
N |
|
AcO |
|
H |
|
|
|
2.28LCAA-CPG
Figure 2.12 LCAA CPG support 2.28 for oligonucleotide SPS.
toward base and allows the use of Fmoc-based chemistry at the 5′-position. A similar linker 2.31, in which the succinyl moiety is replaced by the oxalyl group, is also available (84) and can be cleaved in the final step with 5% methanolic ammonia. The much more robust carbamate linker 2.32 (93) requires concentrated ammonium hydroxide at 56 °C for 2 days to achieve complete release of the target ON.
Four recent examples of universal linkers/supports, in which the first nucleoside is anchored onto the preformed linker–support construct, are shown in Fig. 2.14. The disulfide linker 2.33 has been used to prepare terminal 3′-phosphate ONs (94, 95) through cleavage with a solution of ammonia in dithiothreitol. The photolabile linker 2.34 (96) is used to prepare 3′-alkyl carboxylic acids. The allyl-based linker 2.35 (97) is used to prepare free 3′-OH ONs by cleavage with Pd(0) and treatment with an aqueous buffer at pH 10. The linker 2.36 (98) differs from those discussed so far in
|
|
P1O |
|
O |
NB(P2) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
P1O |
|
|
|
CPG L |
|
|
|
|
|
|
O |
|
NB(P2) |
|
|
O |
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
O |
|
|
|
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
L |
|
O |
|
|
|
|
|
2.29 |
|
|
CPG |
N |
|
|
||
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
O |
Me |
O |
|
|
|
|
|
|
|
|
|
|
2.30 |
|
|
|
|
|
|
|
|
|
|
P1O |
O |
|
|
|
P O |
|
|
|
|
|
|
NB(P2) |
|
|
|
|
|
|
|
|
|
|
||
|
|
1 |
O |
|
|
|
|
|
|
|
|
|
|
NB(P2) |
|
|
|
|
|
||
|
|
O |
|
|
H |
H |
|
|
||
|
|
|
|
|
L |
|
|
|||
|
L |
|
|
|
CPG |
N |
N O |
|
|
|
CPG |
O |
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
O |
|
O |
|
|
|
|
|
O |
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
2.31
2.32
L = LINKER
Figure 2.13 Base-labile linkers 2.29–2.32 for oligonucleotide SPS.

62 SOLID-PHASE SYNTHESIS: OLIGOMERIC MOLECULES
P1O
O  NB(P2)
NB(P2)
O
CPG |
L |
S |
O |
O |
|
O |
S |
P |
|
|
O |
2.33 |
O |
O |
|
|
|
|
|
|
P1O |
|
|
|
O |
|
|
|
OMe |
CPG |
L |
O |
O |
|
|
O |
|
|
|
O |
O |
|
NO2 |
|
2.34 |
|
P1O |
|
|
|
|
O |
NB(P2) |
|
|
|
|
|
O |
O |
|
|
O |
P |
|
L |
O |
O |
|
O |
|
|
|
CPG |
|
|
|
O |
2.35 HN |
OAllyl |
|
O |
|
|
|
N |
N |
|
|
|
N |
|
N |
|
|
 NB(P2)
NB(P2)
P1O
O  NB(P2)
NB(P2)
O O O
P
O O
O N O
|
O |
|
O |
||||||||||
CPG |
L |
|
|
|
|
|
|
|
|
|
|
|
|
(CH2)6 |
N |
||||||||||||
|
|
||||||||||||
O N |
|
||||||||||||
|
|||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|||
|
Me |
||||||||||||
|
|
|
|
|
Me |
||||||||
2.36
Figure 2.14 Universal linkers 2.33–2.36 for oligonucleotide SPS.
