
Neutron Scattering in Biology - Fitter Gutberlet and Katsaras
.pdf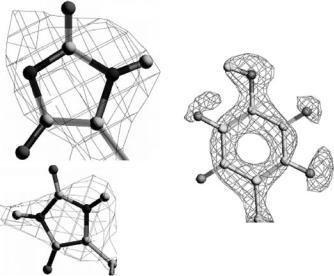
30 C.C. Wilson et al.
His-219:
DD1
NE2
ND1
His-53: DD1
DD2
NE2 ND1
Fig. 2.3. Preliminary density maps from glucose isomerase, measured on PCS at LANSCE (left). The 1.6 ˚A 2Fo-Fc positive nuclear density in blue and the negative nuclear density in red for the side chain of Tyrosine 10 in W3Y rubredoxin (Pf). An example map from neutron data collected at cryogenic temperature 15 K on LADI at the ILL (right)
beamline, promises to deliver a further order of magnitude improvement in performance for neuron protein crystallography.
In addition to its potentially revolutionary applications in biological crystallography, the LADI concept has obvious applications also in the study of smaller molecular systems, and in magnetism. For problems in chemical crystallography, the recently commissioned VIVALDI instrument, sited on a thermal neutron beam line with a wavelength range centered around 1.6 ˚A, is optimal. The image plate approach adopted on LADI is also exploited in the monochromatic Japanese BIX instruments for neutron protein crystallography [8]. Indeed, the BIX concept was the first to use the neutron image plate, developed largely by Niimura [9].
There is also a tremendous opportunity for the exploitation of neutron time-of-flight di raction using single crystal samples. The upgraded SXD instrument recently installed at ISIS has detectors covering over 50% of the solid angle, with a total of 11 PSDs (Fig. 2.2). The detectors are based on the fibre-optically encoded, 3 mm resolution ZnS scintillator detectors previously used on SXD. Future implementations of related instruments, on beamline and moderator choices with better characteristics for larger unit cell structures, will o er the prospect of studying macromolecular structures; as mentioned above, such an instrument is planned for the ISIS Second Target Station, TS2 [10].
2 Neutron Di raction and Protein Crystallography |
31 |
The next generation of high power spallation neutron sources, such as the SNS, being constructed at Oak Ridge National Laboratory in the USA [11] and the spallation source being constructed as part of the J-PARC facility in Tokai, Japan [12] o er new opportunities for neutron protein crystallography. At SNS, a dedicated Macromolecular Neutron Di ractometer (MaNDi) has been designed and is optimized for data collection to 1.5 ˚A resolution from crystals of volume 0.1–1 mm3 with unit cell parameters of 150 ˚A. In addition, the MaNDi instrument will allow neutron data to be collected to between 2.5 and 3 ˚A, on large macromolecular complexes with unit cell dimensions up to 250 to 300 ˚A. The MaNDi instrument will be sited on a decoupled moderator at the 60-Hz SNS source, making optimal use of the SNS design to provide best signal-to-noise and highest possible resolution for large unit cell systems. The ability to measure high resolution neutron di raction data sets within 1–7 days from such large and complex systems promises to greatly extend the range and number of macromolecular systems that are accessible to neutron protein crystallography. It is anticipated that the MaNDi instrument will therefore impact significantly on many areas of structural biology, including enzymology, protein dynamics, drug design, and the study of membrane proteins.
2.2.5 Improvements in Sources
It can be seen from the preceding discussion that the recent successful e orts of instrument designers have led to large improvements in the provision of instrumentation appropriate for the study of macromolecular systems, and it is clear that this will continue. However, a further step function increase in the capabilities of single crystal neutron di raction will be greatly facilitated when the neutron sources themselves are improved. The consequent increases in flux will lead to the study of larger molecules, smaller crystals, and improved variable temperature, and even kinetic studies. For reactors, the prospect for improvement is somewhat limited, given that the power density in the reactor core limits the potential flux increase to around five times that of the present ILL at best. However, it is simple to construct moderators for cold neutrons in reactors and it is also easy to select, guide and detect the longer wavelength neutrons required for biology; there is much prospect for further exploitation of reactor instrumentation in macromolecular single crystal neutron studies.
For pulsed sources, still relatively in their infancy, there is more scope for improvement, already alluded to above. On both the accelerator and target station side, advancing technology allied with increasing experience of operating such sources promise a rich future. There are many moderator options for optimizing wavelength, flux, and pulse width characteristics for instruments on such sources; these issues are being tackled for the first time with the PCS protein crystallography beamline recently constructed at the Los Alamos pulsed source, built on a partially coupled moderator to optimize its high flux performance for biological studies. The ISIS second target station
32 C.C. Wilson et al.
(under construction) also promises a source more optimized for large molecule structural studies than the existing high resolution target station.
On the accelerator side, as mentioned above there are higher intensity sources in construction in the USA (1.4 MW) and in Japan (600 kW). For still higher flux, the present design study for the ESS, the European Spallation Source, aims to provide a 5 MW source with a peak flux of some 30 times that of the present ISIS, with a time averaged flux equivalent to that of the ILL [13]. The use of time-sorted white beams from these sources means that full advantage can be taken of this flux increase and combining this with appropriately optimized moderators should allow for the provision of extremely powerful instrumentation for future single crystal studies, including macromolecular crystallography [14].
Plans are well advanced for instruments optimized for macromolecular crystallography at all of these sources, including a next generation LADI at ILL, the BIX instruments in Japan, MaNDi at SNS and LMX/Proteus at ISIS TS2.
2.3 Information from Neutron Crystallography
Neutron di raction is the method of choice for many crystallographic experiments. Among other characteristics, the nature of the scattering of neutrons by atomic species is such that the method o ers a description of all atoms in a structure at approximately the same level of precision. This “equivalence” of atoms is due to the fact that neutrons are scattered by the nucleus rather than the electrons in an atom, and hence the scattering power does not have the strong dependence on Z found for many other scattering techniques such as X-ray or electron di raction. Of particular relevance to the discussion here is the ability of neutron di raction to detect hydrogen (and deuterium) in chemical and biological structures, where these light elements have approximately equal contributions to the di raction as do the other atoms in the material.
2.3.1 Neutron Crystallography of Molecular Materials
Neutron scattering has played a major role in developing an understanding of how structure a ects the properties of crystalline materials, in areas of relevance to much of modern structural chemistry [15]. Areas accessible to single crystal and powder neutron di raction include organic materials, pharmaceuticals, small biological macromolecules, zeolites, polymer electrolytes, battery materials, catalysts, superconductors, time-resolved and in situ studies, and chemical magnetism [16]. Neutron di raction experiments are often carried out under extreme conditions of sample environment such as high and low temperature, under controlled atmospheres, high pressure and in chemical reaction cells. The combination of X-rays and neutrons is powerful in many
2 Neutron Di raction and Protein Crystallography |
33 |
studies, including the characterization of host–guest interactions in, for example, zeolites, and in determination of charge distributions in crystal structures.
Neutron di raction is unparalleled in its ability to locate hydrogen atoms and refine their positions and thermal parameters. Hydrogen atoms can be located in metal clusters (e.g., hydride ligands) far more reliably than by any other method. Much of the structural work on hydrogen bonded systems (e.g., amino acids, nucleic acid components, carbohydrates, cyclodextrins) has used neutron di raction. In addition, determination of the hydrogen anisotropic displacement parameters in short O–O hydrogen bonds allows, for example, the deduction of the shape of the potential well in which the atom sits. Neutron single crystal di raction has an important role in defining the patterns of “weak” intermolecular interactions in complex molecular and supramolecular structures, as these often crucially involve hydrogen atoms. This leads directly to a strong impact in the expanding area of molecular and crystal engineering. Neutron di raction also gives complementary information to X-ray di raction for charge density studies. In X–N studies the neutron parameters fix the nuclear positions and the X-ray data determine the electron density involved in bonding and nonbonding interactions.
2.3.2 Neutron Crystallography in Structural Biology
In addition to the major impact the technique has had in chemical crystallography, single crystal neutron di raction has made a significant and important contribution in the determination of biologically important structures. There are a number of examples where single crystal studies of proteins have had a profound influence on our understanding of how the protein might function. An excellent summary of the application of single crystal neutron di raction in the biological area was given by Knott and Schoenborn [17] and in a more recent review by Tsyba and Bau [18].
In the field of structural biology, the relation between structure and function has been established since the early days of protein structure determination. A full understanding of this relationship depends on an appreciation of the detailed molecular interactions involved. These interactions occur through mechanisms such as hydrogen bonding, charge transfer, and other nonbonded interactions, and many of these are governed by the location of hydrogen atoms. Accurate neutron di raction studies can define to high precision the geometry of an active site, and the role which this may play in important interactions, for example with drug molecules. Hydrogen atoms inevitably decorate much of the outer regions of both protein molecules and interacting small molecules. Many important protein functions can thus depend on the presence or absence of just one hydrogen atom and it is clearly important to locate these accurately.
It is this crucial aspect of the role of hydrogen atoms in biological function that has led to the continued pursuit of routine neutron protein crystallography through many years of e ort and in spite of the intrinsic di culty of the
34 C.C. Wilson et al.
experiments. There are other areas where single crystal neutron di raction has a unique contribution to make in this field:
–The ability of neutron scattering to distinguish clearly between nitrogen, carbon, and oxygen is important, for example in determining the orientation of histidine, apargine, and glutamine.
–The very large scattering length di erence (“contrast”) between hydrogen and deuterium can be exploited to allow the determination of exchangeable hydrogen atoms, yielding information on protein dynamics and on solvent accessibility.
–In a traditionally powerful area of application of neutron single crystal studies, analysis of thermal motions of hydrogen-containing groups in the structure can give information on the physics underlying the structure.
–Protein–solvent interactions are critical to life. The ordered solvent structure around protein molecules has also been elucidated by high resolution neutron single crystal di raction studies. Neutron studies can reveal both the position and the orientation of the water molecules by locating not only oxygen atom positions but also the hydrogen (or deuterium) atoms as well.
2.3.3 Sample and Data Requirements for Single Crystal Neutron Di raction
The requirement for rather large protein single crystals is one of the main drawbacks of single crystal neutron di raction compared with X-ray methods. With the relatively low flux of neutron sources and the rather weak scattering of most materials, usually crystals of several cubic millimeters are required to allow collection of a good data set in a reasonable data collection time. The limit is usually regarded as being around 1 mm3, given that data collection times of greater than 10–14 days per data set are usually impractical. This limit is now being lowered at facilities such as the LADI instrument at ILL and there are significant e orts being made by source and instrument designers to allow it to be further reduced. The major limitation on the signal-to-noise ratio in neutron di raction from biological (as well as all other hydrogenous materials) is that hydrogen has a large incoherent neutron scattering factor of 80 barns that produces a high level background that significantly reduces the signal to noise ratio of the di raction data. The corresponding value for the deuterium isotope is 2 barns; isotopic substitution of deuterium for hydrogen therefore results in huge reductions in the incoherent scattering background and order of magnitude improvement in signal to noise. It is now possible to prepare such isotopically H/D substituted or labeled proteins and other macromolecules in the laboratory. The ability to clone and over-express target proteins of interest for biochemical and biophysical analysis in host microbial systems is now considered routine in laboratories world-wide. These similar microbial systems can be adapted to growth in heavy water (D2O) solutions and when fed with deuterated carbon sources, are able to produce functional
2 Neutron Di raction and Protein Crystallography |
35 |
protein molecules in which all hydrogen atoms in the structure have been replaced by the deuterium [19]. The reduced incoherent scattering background from crystals of such deuterated proteins results in order of magnitude improvements in signal to noise. This is a critical advantage for neutron protein crystallography that, together with instrument developments, promises to deliver further 100-fold reductions in the size of sample than can be used on future instruments, making larger and more complex systems amenable to neutron protein structure determination.
2.4 Brief Review of the Use of Neutron Di raction in the Study of Biological Structures
Much of biological structure and function is mediated by hydrogen bonding interactions and proper account of the hydrogen atom structure of biological systems brings understanding of biological process at the most fundamental level. Neutron protein crystallography can play a key role in structural molecular biology by locating hydrogen atoms and water positions in proteins that cannot be seen by X-ray analysis alone. While NMR and X-ray techniques have unrivalled capacity for high-throughput structure determination, neutron di raction, and other specialized techniques, make key and unique contributions to the field. Hydrogen bonding interactions mediate most of biological structure and function and the location of even a single hydrogen atom can help determine and define the mechanism and pathway of complex biological processes. However, hydrogen atoms can only be seen (if at all) by X-rays if protein crystals are su ciently well ordered to provide data to atomic resolutions (<1.1 ˚A). While the position of many hydrogen atoms can be reliably inferred from the chemical groups to which they are bound, the positions of other more labile – and perhaps more interesting – atoms cannot and must be determined by other techniques.
A number of structural studies have been carried out using neutron diffraction, focusing very much on structures where the X-ray work has left some ambiguity over an important aspect of hydrogen atom location or solvent structure. In most cases, the neutron data have provided an indication of some structural feature which was left undefined by the X-ray data. Often neutron di erence Fourier maps based on the known X-ray structure can reveal incorrect assignment of atoms or misorientation of important groups. For example, positions of oxygen and nitrogen atoms in asparagine and glutamine side-chain amides were distinguished and switched based on information from neutron data. Joint X-ray and neutron refinements were used to study the structure of ribonuclease A [20]. Neutron data extending to around 2 ˚A resolution were collected from a 30 mm3 sample at the NBS reactor at NIST. The neutron structure refinement showed the proper rotation of key amino groups, including the orientation of the four histidine side chains, the Arg39 side chain was also completely reoriented, and the catalytically important Lys41 was completely rebuilt based on the neutron Fourier maps. The
36 C.C. Wilson et al.
new configuration of this group increased the likelihood that it was involved in the activity of this molecule. The solvent structure was also significantly modified in the joint refinement. High resolution studies have also allowed high levels of detail to be obtained in smaller, and therefore well ordered, protein structures such as lysozyme [21, 22] and crambin [23].
However, such studies have always been performed in the limiting context of the need for very large crystals, which has severely limited the number and range of structures amenable for study. Even at the most intense neutron sources, such as the 58 MW high flux reactor at ILL, conventional instruments such as D19 that is prominent in neutron chemical crystallography, have been restricted to small unit cell biological systems, such as vitamin B12 [25, 26], lysozyme [21] and to fibre di raction of biological polymers, hampered by the limited capacity of available instrumentation (see Forsyth et al. this volume). The new generation of protein crystallography instruments, such as LADI at ILL, the BIX instruments at JAERI and PCS at LANSCE, have delivered order of magnitude gains in performance that make feasible studies of larger biological complexes and smaller crystals than was previously possible. The further order of magnitude improvements in performance provided by the protein crystallography instruments planned at the 1.4 MW SNS and the 0.6 MW J-SNS facilities will be decisive in opening new fields of research.
2.4.1 Location of Hydrogen Atoms
The transfer and exchange of hydrogen atoms in biological systems is of fundamental importance in biology. The determination of the hydration and protonation state of proteins is thus a major focus in neutron protein crystallography, particularly where the geometry of these cannot reliably be predicted on the basis of the X-ray structure. A fine early example of this was given in the study of vitamin B12 [24]. The continuing interest in vitamin B12 coenzyme, both for interest in itself and as a model system for large molecule studies, has led to repeated high resolution neutron single crystal studies being carried out at both room and low temperature. Refinement against room temperature data showed that there was significant reorientation of part of the structure compared with the earlier X-ray study, due to a redistribution of hydrogen bonds around the phosphate group [25]. There is a complex hydrogen bonding network, made still more complex by the presence of disorder in one of the side chains of the corrin ring, characterized by the neutron diffraction studies. The low temperature study [26], carried out at 15 K, showed a substantial reduction in the static disorder present.
The location of a single hydrogen atom can also have profound implications for the biological mechanism of a large molecule, for example in the oxygen carrying mechanism of haemoglobin. The local geometry around the heme group was also the motivation for the early neutron di raction
2 Neutron Di raction and Protein Crystallography |
37 |
studies on myoglobin [27]. Subsequent work on this system allowed an important distinction to be made between the binding modes of CO and O2 molecules, with the finding that the imidazole group of His-64 was not protonated on CO binding [28], while it was found to be so in the situation of O2 binding [29].
Another case in which the presence or absence of a single hydrogen atom has implications for the function of a protein is in the mechanism of action of the serine proteases [30]. Specifically, in the case of trypsin the location of a single proton, either on Histidine-57 or Asparagine-102, has implications for the catalytic activity of this protein. The neutron single crystal di raction, collected to 2.2 ˚A resolution at Brookhaven on a 1.5 mm3 crystal which had been soaked in D2O to replace most of the exchangeable H for D, showed that the proton was attached to His-57, showing this to be the chemical base in the hydrolysis reaction [31]. This meant that mechanisms for the action of the serine proteases which had Asp-102 as the base could be eliminated.
In recent key studies, H/D locations determined at medium resolution ( 2 ˚A) by neutron protein crystallography have provided additional information that could not be determined from atomic resolution (<1.2 ˚A) synchrotron X-ray data alone. For example, the 2 ˚A neutron structure of an aspartic protease, endothiapepsin, a transition state analogue complex, directly revealed the key hydrogen positions at the catalytic site of the protein (Fig. 2.4). The data provide convincing evidence that Asp-215 is protonated
Endothiapepsin at 2.1 Å
Asp-215
Asp-32
Fig. 2.4. The 2 ˚A neutron structure of Endothiapepsin directly revealed the key hydrogen positions at the catalytic site of the protein
38 C.C. Wilson et al.
and that Asp-32 is the negatively charged residue in the transition state complex. This has an important bearing on mechanistic proposals for this class of proteinase, resolving the long-standing controversy over the catalytic mechanism in this important family of enzymes [32]. In myoglobin, where hydrogen atom positions could not be visualized from X-ray data at <1.15 ˚A [33], the neutron structure of the perdeuterated protein determined at 2.0 ˚A resolution enabled deuterium atom positions of interest to be determined [29]. In particular, this work allowed the determination of the protonation sites as positive peaks near the Nε or Nδ atoms of all 12 histidines in Mb at pH 6.2.
As for smaller organic structures, the hydrogen atoms in terminal methyl groups in proteins undergo torsional or rotational motion. The orientations of these “rotor” methyl groups are di cult to determine, but neutron di raction has had success in doing so. One example is in the structure of trypsin, where the examination of neutron scattering densities as a function of torsion angle for the rotation of these clearly revealed the correct orientation and hence the location of the hydrogen atoms [34].
2.4.2 Solvent Structure
Neutron di raction is also ideally suited to the study of solvent structure in biological systems. Typically 40–60% of an average protein crystal actually consists of water. The relative scattering power of solvent water is larger for neutron di raction than X-ray di raction, especially so in the case of heavy (D2O) water and in high resolution studies, so that neutron analysis also gives the opportunity of distinguishing the proton/deuterium positions in the solvent structure. This has led, for example, to a detailed description of the hydration structure in vitamin B12 [35]. There has also been success in studying the solvent structure in larger molecule systems, especially in the case where water molecules are well ordered, such as when they are directly bound to the protein surface [36]. The interaction of other solvents with proteins can also be investigated, for example an analysis of the interaction of dimethyl sulphoxide (DMSO) with lysozyme [21] showed that the DMSO molecules interact with the protein surface both through hydrogen bonds and bonding through solvent methyl groups to the hydrophobic parts of the protein molecule, without significantly changing the protein configuration. Such studies o er the possibility of resolving the precise nature of the solvent–protein and solvent–solvent interactions present. This potential has recently begun to be more fully explored by combining high resolution studies with advanced modeling of solvent structure [37]. In a recent technical advance, it has been shown that by collecting neutron data at cryo temperatures, the dynamic disorder within a protein crystal is reduced, the definition of the nuclear density is improved, and a comparison between the 15 and 293 K neutron structures shows that overall, twice as many bound waters (as D2O) are identified at 15 K than at 293 K [38].
2 Neutron Di raction and Protein Crystallography |
39 |
2.4.3 Hydrogen Exchange
While NMR and radioactive labeling are the most common tools for monitoring hydrogen exchange in proteins, the high contrast between H and D makes neutron di raction a realistic alternative [39]. In particular, neutron di raction provides a powerful method of monitoring the propensity for exchange of amide protons in even large protein structures. The method does not of course provide kinetic data on hydrogen exchange, but instead provides snapshots of the structure. Such snapshots, however, are su cient to indicate the degree of exchange and hence to highlight the less flexible regions of the protein molecule. For example, in a series of experiments conducted at the BIX-3 instrument at JAERI on the small and unusually thermostable protein rubredoxin from P. furiosus (an organism that grows optimally at 373 K), comparison of the hydrogen-bonding patterns of partially deuterated wild-type and triple mutants proteins provided insight into the H/D-exchange pattern of the N–H amide bonds of the protein backbone and information on the mechanism of unfolding [40].
2.4.4 Low Resolution Studies
In the presence of disorder, or where extremely large structural components are to be resolved, lower resolution data are of value because they reflect measurements on a scale (of order 10 ˚A) where the scattering density of the various components is roughly constant. Such lower resolution studies can be carried out with neutrons of wavelength of the order of 7–8 ˚A, thus benefitting from the higher reflectivity for scattering of such neutrons and allowing smaller crystals to be used. In the case of very large biological molecular complexes, or in cases where a major component of the structure is disordered, such studies can provide information on the location of individual components in large and complex systems, such as lipid, detergent, carbohydrate, or nucleic acids, which can be essential to construct a biologically meaningful picture of the structure as a whole.
Low resolution studies, usually coupled with contrast variation, have been used to obtain information on membrane-bound proteins [41], and to see lipids associated with protein complexes and assemblies [42], and in one-dimensional di raction studies of membrane structures themselves [43]. In a recent example of work on membrane protein structure the detergent structure present in crystals of the peripheral light-harvesting complex of the purple bacteria Rhodopseudomonas acidophila has been determined at a maximal resolution of 12 ˚A by neutron crystallography (Fig. 2.5) [44].
2.4.5 Other Biologically Relevant Molecules
Neutrons can also be used to study other biologically relevant structures, such as pharmaceuticals and other biologically active molecules. These are in
